PWMs for MOUSE transcription factors
| Model | LOGO | Transcription factor | Model length | Quality | Consensus | Origin | wAUC | Best AUC | Benchmark datasets | Aligned words | TF family | TF subfamily | MGI | EntrezGene | UniProt ID | UniProt AC | Comment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHR_MOUSE.H10MO.B |  |
Ahr | 9 | B | dKhGCGTGh | HOCOMOCO v9 | 157 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 105043 | 11622 | AHR_MOUSE | P30561 | ||||
| AIRE_MOUSE.H10MO.C |  |
Aire | 18 | C | hnnGGWWnddWWGGdbWh | HOCOMOCO v9 | 41 | AIRE{5.3.1} | AIRE{5.3.1.0.1} | 1338803 | 11634 | AIRE_MOUSE | Q9Z0E3 | ||||
| ALX1_MOUSE.H10MO.B |  |
Alx1 | 12 | B | vTRATYGnATTA | HOCOMOCO v9 | 33 | Paired-related HD factors{3.1.3} | ALX{3.1.3.1} | 104621 | 216285 | ALX1_MOUSE | Q8C8B0 | ||||
| ANDR_MOUSE.H10MO.B | 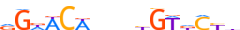 |
Ar | 16 | B | RGWWCWbndhGTdCYd | HOCOMOCO v9 | 131 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 88064 | 11835 | ANDR_MOUSE | P19091 | ||||
| AP2A_MOUSE.H10MO.A |  |
Tfap2a | 9 | A | SCCYGvGGS | HOCOMOCO v9 | 1952 | AP-2{1.3.1} | AP-2alpha{1.3.1.0.1} | 104671 | 21418 | AP2A_MOUSE | P34056 | ||||
| AP2B_MOUSE.H10MO.B |  |
Tfap2b | 10 | B | GSCYGRGGRv | HOCOMOCO v9 | 30 | AP-2{1.3.1} | AP-2beta{1.3.1.0.2} | 104672 | 21419 | AP2B_MOUSE | Q61313 | ||||
| AP2C_MOUSE.H10MO.A |  |
Tfap2c | 9 | A | SCCYGvGGS | HOCOMOCO v9 | 1615 | AP-2{1.3.1} | AP-2gamma{1.3.1.0.3} | 106032 | 21420 | AP2C_MOUSE | Q61312 | ||||
| AP2D_MOUSE.H10MO.D |  |
Tfap2d | 14 | D | nGbCCGRGGCnCGY | HOCOMOCO v9 | 5 | AP-2{1.3.1} | AP-2delta{1.3.1.0.4} | 2153466 | 226896 | AP2D_MOUSE | Q91ZK0 | ||||
| ARI3A_MOUSE.H10MO.D |  |
Arid3a | 22 | D | dWTTAAWYvRdWWYAAAWYWWW | HOCOMOCO v9 | 44 | ARID-related factors{3.7.1} | ARID3{3.7.1.3} | 1328360 | 13496 | ARI3A_MOUSE | Q62431 | ||||
| ARI3A_MOUSE.H10MO.S |  |
Arid3a | 7 | S | RATTAAA | HOCOMOCO v9 | 53 | ARID-related factors{3.7.1} | ARID3{3.7.1.3} | 1328360 | 13496 | ARI3A_MOUSE | Q62431 | Secondary motif. | |||
| ARI5B_MOUSE.H10MO.C |  |
Arid5b | 13 | C | nbYKnRTATTSKd | HOCOMOCO v9 | 19 | ARID-related factors{3.7.1} | ARID5{3.7.1.5} | 2175912 | 71371 | ARI5B_MOUSE | Q8BM75 | ||||
| ARNT2_MOUSE.H10MO.D |  |
Arnt2 | 12 | D | RnSGTGGvRSRS | HOCOMOCO v9 | 18 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 107188 | 11864 | ARNT2_MOUSE | Q61324 | ||||
| ARNT_MOUSE.H10MO.B |  |
Arnt | 8 | B | bYACGTGh | HOCOMOCO v9 | 283 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 88071 | 11863 | ARNT_MOUSE | P53762 | ||||
| ATF1_MOUSE.H10MO.B |  |
Atf1 | 10 | B | nTGACGTSMv | HOCOMOCO v9 | 43 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 1298366 | 11908 | ATF1_MOUSE | P81269 | ||||
| ATF2_MOUSE.H10MO.B |  |
Atf2 | 9 | B | vTGAYGThM | HOCOMOCO v9 | 109 | Jun-related factors{1.1.1} | ATF-2-like factors{1.1.1.3} | 109349 | 102641666; 11909 | ATF2_MOUSE | P16951 | Model is applicable to several TFs or complex subunits: ATF2_MOUSE, ATF4_MOUSE. | |||
| ATF3_MOUSE.H10MO.A |  |
Atf3 | 9 | A | vTGAYKYvd | HOCOMOCO v9 | 2096 | Fos-related factors{1.1.2} | ATF-3-like factors{1.1.2.2} | 109384 | 11910 | ATF3_MOUSE | Q60765 | ||||
| ATOH1_MOUSE.H10MO.C |  |
Atoh1 | 11 | C | RRCAGMTGKbv | ChIP-Seq | 0.8927500766335188 | 0.8927500766335188 | 1 | 502 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 104654 | 11921 | ATOH1_MOUSE | P48985 | |
| BACH1_MOUSE.H10MO.C |  |
Bach1 | 14 | C | YGCTGAGTCAYGST | HOCOMOCO v9 | 8 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 894680 | 12013 | BACH1_MOUSE | P97302 | ||||
| BARX2_MOUSE.H10MO.D |  |
Barx2 | 10 | D | hYnWTAATKR | HOCOMOCO v9 | 14 | NK-related factors{3.1.2} | BARX{3.1.2.2} | 109617 | 12023 | BARX2_MOUSE | O08686 | ||||
| BATF_MOUSE.H10MO.B |  |
Batf | 10 | B | vTGAGTCAbh | HOCOMOCO v9 | 1924 | B-ATF-related factors{1.1.4} | B-ATF{1.1.4.0.1} | 1859147 | 53314 | BATF_MOUSE | O35284 | ||||
| BCL6_MOUSE.H10MO.C |  |
Bcl6 | 13 | C | dTYCCTRGAvRKh | ChIP-Seq | 0.728609363450918 | 0.7708335218509506 | 3 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | BCL6 factors{2.3.3.22} | 107187 | 12053 | BCL6_MOUSE | P41183 | |
| BHE40_MOUSE.H10MO.B |  |
Bhlhe40 | 9 | B | KCACGTGMb | HOCOMOCO v9 | 0.8828224771457921 | 0.8828224771457921 | 1 | 2327 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 1097714 | 20893 | BHE40_MOUSE | O35185 | |
| BHE41_MOUSE.H10MO.D |  |
Bhlhe41 | 20 | D | dCnRnSTCRMGTGMhKRnRn | HOCOMOCO v9 | 10 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 1930704 | 79362 | BHE41_MOUSE | Q99PV5 | ||||
| BMAL1_MOUSE.H10MO.C |  |
Arntl | 14 | C | dGvWCACGTGAChS | HOCOMOCO v9 | 15 | PAS domain factors{1.2.5} | Arnt-like factors{1.2.5.2} | 1096381 | 11865 | BMAL1_MOUSE | Q9WTL8 | ||||
| BRAC_MOUSE.H10MO.D | 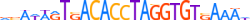 |
T | 25 | D | vSAWARTSACACCTAGGTGTSAAAW | HOCOMOCO v9 | 26 | Brachyury-related factors{6.5.1} | T (Brachyury){6.5.1.0.1} | 98472 | 20997 | BRAC_MOUSE | P20293 | ||||
| BRCA1_MOUSE.H10MO.D |  |
Brca1 | 9 | D | bKKbKGTTG | HOCOMOCO v9 | 72 | 104537 | 12189 | BRCA1_MOUSE | P48754 | ||||||
| CDC5L_MOUSE.H10MO.D |  |
Cdc5l | 15 | D | vbGnKdTAAYRWAWb | HOCOMOCO v9 | 10 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 1918952 | 71702 | CDC5L_MOUSE | Q6A068 | ||||
| CDX1_MOUSE.H10MO.C |  |
Cdx1 | 8 | C | hTTKAWGd | HOCOMOCO v9 | 30 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 88360 | 12590 | CDX1_MOUSE | P18111 | ||||
| CDX2_MOUSE.H10MO.C |  |
Cdx2 | 8 | C | WTTTAYdd | HOCOMOCO v9 | 36 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 88361 | 12591 | CDX2_MOUSE | P43241 | ||||
| CDX4_MOUSE.H10MO.A |  |
Cdx4 | 12 | A | WRATTTAYdRbh | ChIP-Seq | 0.8513936928849216 | 0.8700411514211168 | 2 | 500 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 88362 | 12592 | CDX4_MOUSE | Q07424 | |
| CEBPA_MOUSE.H10MO.B |  |
Cebpa | 12 | B | dvTTKdGMAAYh | ChIP-Seq | 0.8181687579641503 | 0.8839223541807075 | 2 | 501 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 99480 | 12606 | CEBPA_MOUSE | P53566 | |
| CEBPB_MOUSE.H10MO.A |  |
Cebpb | 11 | A | dRTTGCRYAAY | ChIP-Seq | 0.9176046304239921 | 0.9351823065612739 | 10 | 505 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 88373 | 12608 | CEBPB_MOUSE | P28033 | |
| CEBPD_MOUSE.H10MO.B | 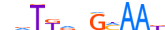 |
Cebpd | 11 | B | dvTKdbGMAAK | HOCOMOCO v9 | 79 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 103573 | 12609 | CEBPD_MOUSE | Q00322 | ||||
| CEBPE_MOUSE.H10MO.A |  |
Cebpe | 12 | A | dTKSGMAAThdb | HOCOMOCO v9 | 803 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 103572 | 110794 | CEBPE_MOUSE | Q6PZD9 | ||||
| CEBPG_MOUSE.H10MO.C |  |
Cebpg | 13 | C | MTKKbGYAATbKb | HOCOMOCO v9 | 23 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 104982 | 12611 | CEBPG_MOUSE | P53568 | ||||
| CEBPZ_MOUSE.H10MO.D |  |
Cebpz | 11 | D | dSTSATTGGCT | HOCOMOCO v9 | 28 | 109386 | 12607 | CEBPZ_MOUSE | P53569 | ||||||
| COE1_MOUSE.H10MO.C |  |
Ebf1 | 13 | C | bYCCCWKGGGRvh | ChIP-Seq | 0.9285894408954788 | 0.9285894408954788 | 1 | 503 | Early B-Cell Factor-related factors{6.1.5} | EBF1 (COE1){6.1.5.0.1} | 95275 | 13591 | COE1_MOUSE | Q07802 | |
| COT1_MOUSE.H10MO.B |  |
Nr2f1 | 9 | B | YWRAGGTCA | HOCOMOCO v9 | 86 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352451 | 13865 | COT1_MOUSE | Q60632 | ||||
| COT1_MOUSE.H10MO.S |  |
Nr2f1 | 12 | S | SdbYWRAGKTCA | HOCOMOCO v9 | 84 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352451 | 13865 | COT1_MOUSE | Q60632 | Secondary motif. | |||
| COT2_MOUSE.H10MO.B |  |
Nr2f2 | 10 | B | MWRAGGTCAd | HOCOMOCO v9 | 79 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352452 | 11819 | COT2_MOUSE | P43135 | ||||
| COT2_MOUSE.H10MO.S |  |
Nr2f2 | 15 | S | vRGKbCAdRGRKYvv | HOCOMOCO v9 | 82 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352452 | 11819 | COT2_MOUSE | P43135 | Secondary motif. | |||
| CREB1_MOUSE.H10MO.B |  |
Creb1 | 9 | B | vTGACGYvd | HOCOMOCO v9 | 308 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 88494 | 12912 | CREB1_MOUSE | Q01147 | ||||
| CREM_MOUSE.H10MO.C | 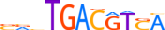 |
Crem | 11 | C | SRvTGACGTSA | HOCOMOCO v9 | 30 | CREB-related factors{1.1.7} | CREB-like factors{1.1.7.1} | 88495 | 12916 | CREM_MOUSE | P27699 | ||||
| CRX_MOUSE.H10MO.A |  |
Crx | 12 | A | hdRdvRGATTAd | ChIP-Seq | 0.8561267887670638 | 0.8629600802653843 | 2 | 500 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 1194883 | 12951 | CRX_MOUSE | O54751 | |
| CRX_MOUSE.H10MO.S |  |
Crx | 8 | S | vdGATTAd | HOCOMOCO v9 | 0.8188379527525089 | 0.8201527275822534 | 2 | 53 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 1194883 | 12951 | CRX_MOUSE | O54751 | Secondary motif. |
| CTCF_MOUSE.H10MO.A |  |
Ctcf | 20 | A | hbRCCRShAGGKGGCGSbvb | ChIP-Seq | 0.9771765471040378 | 0.9973538813149299 | 35 | 500 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 109447 | 13018 | CTCF_MOUSE | Q61164 | |
| CUX1_MOUSE.H10MO.C |  |
Cux1 | 14 | C | vbRvndATYRRTbb | HOCOMOCO v9 | 97 | HD-CUT factors{3.1.9} | CUX{3.1.9.2} | 88568 | 13047 | CUX1_MOUSE | P53564 | ||||
| CXXC1_MOUSE.H10MO.D |  |
Cxxc1 | 7 | D | YGKTKKY | HOCOMOCO v9 | 63 | 1921572 | 74322 | CXXC1_MOUSE | Q9CWW7 | ||||||
| DBP_MOUSE.H10MO.B |  |
Dbp | 11 | B | vTKRYGTAAbv | HOCOMOCO v9 | 26 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 94866 | 13170 | DBP_MOUSE | Q60925 | ||||
| DDIT3_MOUSE.H10MO.C |  |
Ddit3 | 13 | C | RRvKATTGCAbbb | HOCOMOCO v9 | 42 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 109247 | 13198 | DDIT3_MOUSE | P35639 | ||||
| DLX2_MOUSE.H10MO.D |  |
Dlx2 | 8 | D | ATAATTAn | HOCOMOCO v9 | 6 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 94902 | 13392 | DLX2_MOUSE | P40764 | ||||
| DLX3_MOUSE.H10MO.C |  |
Dlx3 | 10 | C | RMTAATTRvh | HOCOMOCO v9 | 35 | NK-related factors{3.1.2} | DLX{3.1.2.5} | 94903 | 13393 | DLX3_MOUSE | Q64205 | ||||
| E2F1_MOUSE.H10MO.A |  |
E2f1 | 14 | A | ndndGGCGSSRRRn | HOCOMOCO v9 | 2136 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 101941 | 13555 | E2F1_MOUSE | Q61501 | ||||
| E2F2_MOUSE.H10MO.B |  |
E2f2 | 10 | B | RGCGCGMAWC | HOCOMOCO v9 | 8 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 1096341 | 242705 | E2F2_MOUSE | P56931 | ||||
| E2F3_MOUSE.H10MO.B |  |
E2f3 | 10 | B | SGCGCGRAAn | HOCOMOCO v9 | 12 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 1096340 | 13557 | E2F3_MOUSE | O35261 | ||||
| E2F4_MOUSE.H10MO.C |  |
E2f4 | 12 | C | vddGGCGGGRRv | ChIP-Seq | 0.7434912563099131 | 0.77619074877172 | 4 | 500 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 103012 | 104394 | E2F4_MOUSE | Q8R0K9 | |
| E2F5_MOUSE.H10MO.B |  |
E2f5 | 10 | B | nGCGCCAAAh | HOCOMOCO v9 | 12 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 105091 | 13559 | E2F5_MOUSE | Q61502 | ||||
| E2F6_MOUSE.H10MO.A |  |
E2f6 | 12 | A | ndRGCGGGARvv | HOCOMOCO v9 | 2029 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 1354159 | 50496 | E2F6_MOUSE | O54917 | ||||
| E2F7_MOUSE.H10MO.D |  |
E2f7 | 13 | D | nAdGGCGCGAAAn | HOCOMOCO v9 | 5 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 1289147 | 52679 | E2F7_MOUSE | Q6S7F2 | ||||
| E4F1_MOUSE.H10MO.D |  |
E4f1 | 9 | D | nGTGACGTM | HOCOMOCO v9 | 9 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 109530 | 13560 | E4F1_MOUSE | Q8CCE9 | ||||
| EGR1_MOUSE.H10MO.A |  |
Egr1 | 11 | A | nGCGKGGGYGK | HOCOMOCO v9 | 2104 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 95295 | 13653 | EGR1_MOUSE | P08046 | ||||
| EGR2_MOUSE.H10MO.C |  |
Egr2 | 11 | C | dGvGKGGGCGK | HOCOMOCO v9 | 23 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 95296 | 13654 | EGR2_MOUSE | P08152 | ||||
| EGR3_MOUSE.H10MO.D |  |
Egr3 | 11 | D | WRMGTGGGTGb | HOCOMOCO v9 | 8 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 1306780 | 13655 | EGR3_MOUSE | P43300 | ||||
| EGR4_MOUSE.H10MO.D |  |
Egr4 | 11 | D | GGSGGYAGGGM | HOCOMOCO v9 | 6 | Three-zinc finger Krüppel-related factors{2.3.1} | EGR factors{2.3.1.3} | 99252 | 13656 | EGR4_MOUSE | Q9WUF2 | ||||
| EHF_MOUSE.H10MO.A |  |
Ehf | 13 | A | hWASCCGGAARTR | HOCOMOCO v9 | 313 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 1270840 | 13661 | EHF_MOUSE | O70273 | ||||
| ELF1_MOUSE.H10MO.B |  |
Elf1 | 10 | B | vSCGGAARYd | HOCOMOCO v9 | 76 | Ets-related factors{3.5.2} | Elf-1-like factors{3.5.2.3} | 107180 | 13709 | ELF1_MOUSE | Q60775 | ||||
| ELF2_MOUSE.H10MO.C |  |
Elf2 | 15 | C | YRnCAGGAAGbGnnK | HOCOMOCO v9 | 9 | Ets-related factors{3.5.2} | Elf-1-like factors{3.5.2.3} | 1916507 | 69257 | ELF2_MOUSE | Q9JHC9 | ||||
| ELF3_MOUSE.H10MO.D |  |
Elf3 | 14 | D | dRSdWnCAGGAART | HOCOMOCO v9 | 14 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 1101781 | 13710 | ELF3_MOUSE | Q3UPW2 | ||||
| ELF5_MOUSE.H10MO.D |  |
Elf5 | 11 | D | WhvMGGAARYd | HOCOMOCO v9 | 39 | Ets-related factors{3.5.2} | EHF-like factors{3.5.2.4} | 1335079 | 13711 | ELF5_MOUSE | Q8VDK3 | ||||
| ELK1_MOUSE.H10MO.B |  |
Elk1 | 9 | B | vMMGGAARY | HOCOMOCO v9 | 160 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 101833 | 13712 | ELK1_MOUSE | P41969 | ||||
| ELK3_MOUSE.H10MO.D |  |
Elk3 | 12 | D | vnCMGGAWGTGC | HOCOMOCO v9 | 7 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 101762 | 13713 | ELK3_MOUSE | P41971 | ||||
| ELK4_MOUSE.H10MO.B |  |
Elk4 | 11 | B | vWCCGGAAGYv | HOCOMOCO v9 | 54 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 102853 | 13714 | ELK4_MOUSE | P41158 | ||||
| EPAS1_MOUSE.H10MO.D |  |
Epas1 | 13 | D | GKGnGTASGTSKG | HOCOMOCO v9 | 13 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 109169 | 13819 | EPAS1_MOUSE | P97481 | ||||
| ERG_MOUSE.H10MO.A |  |
Erg | 11 | A | RCCGGAARTSv | HOCOMOCO v9 | 559 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95415 | 13876 | ERG_MOUSE | P81270 | ||||
| ERR1_MOUSE.H10MO.A |  |
Esrra | 9 | A | hMRAGGKCA | HOCOMOCO v9 | 2051 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1346831 | 26379 | ERR1_MOUSE | O08580 | ||||
| ERR2_MOUSE.H10MO.B |  |
Esrrb | 9 | B | bSAAGGTCA | HOCOMOCO v9 | 0.9533452485355083 | 0.9533452485355083 | 1 | 3687 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1346832 | 26380 | ERR2_MOUSE | Q61539 | |
| ERR3_MOUSE.H10MO.B |  |
Esrrg | 9 | B | TYAAGGTCA | HOCOMOCO v9 | 14 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1347056 | 26381 | ERR3_MOUSE | P62509 | ||||
| ESR1_MOUSE.H10MO.B |  |
Esr1 | 18 | B | RRGTCAbvbYSvCCYbvv | HOCOMOCO v9 | 568 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1352467 | 13982 | ESR1_MOUSE | P19785 | ||||
| ESR2_MOUSE.H10MO.B |  |
Esr2 | 15 | B | RGGTCAbdbYRvYYY | HOCOMOCO v9 | 382 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 109392 | 13983 | ESR2_MOUSE | O08537 | ||||
| ESR2_MOUSE.H10MO.S |  |
Esr2 | 7 | S | RGGTCAv | HOCOMOCO v9 | 393 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 109392 | 13983 | ESR2_MOUSE | O08537 | Secondary motif. | |||
| ETS1_MOUSE.H10MO.B |  |
Ets1 | 9 | B | dSMGGAWRY | HOCOMOCO v9 | 0.7011989312027058 | 0.7011989312027058 | 1 | 151 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95455 | 23871 | ETS1_MOUSE | P27577 | |
| ETS2_MOUSE.H10MO.C |  |
Ets2 | 11 | C | vhvGGARRhdn | HOCOMOCO v9 | 42 | 95456 | 23872 | ETS2_MOUSE | P15037 | ||||||
| ETV4_MOUSE.H10MO.B |  |
Etv4 | 8 | B | vMGGAAGY | HOCOMOCO v9 | 47 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 99423 | 18612 | ETV4_MOUSE | P28322 | ||||
| ETV5_MOUSE.H10MO.D |  |
Etv5 | 13 | D | SMSMGGAdSdWMY | HOCOMOCO v9 | 15 | Ets-related factors{3.5.2} | Elk-like factors{3.5.2.2} | 1096867 | 104156 | ETV5_MOUSE | Q9CXC9 | ||||
| EVI1_MOUSE.H10MO.B |  |
Mecom | 16 | B | dWGAYAAGATAAnMbd | HOCOMOCO v9 | 141 | Factors with multiple dispersed zinc fingers{2.3.4} | Evi-1-like factors{2.3.4.14} | 95457 | EVI1_MOUSE | P14404 | |||||
| EVX1_MOUSE.H10MO.C | 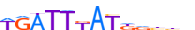 |
Evx1 | 13 | C | WGATTKATKdbhh | ChIP-Seq | 0.8192213945259845 | 0.8192213945259846 | 1 | 500 | HOX-related factors{3.1.1} | EVX (Even-skipped homolog){3.1.1.10} | 95461 | 14028 | EVX1_MOUSE | P23683 | |
| EVX2_MOUSE.H10MO.C |  |
Evx2 | 12 | C | bTTTATKRYhbb | ChIP-Seq | 0.9207906773922094 | 0.9207906773922094 | 1 | 505 | HOX-related factors{3.1.1} | EVX (Even-skipped homolog){3.1.1.10} | 95462 | 14029 | EVX2_MOUSE | P49749 | |
| FEV_MOUSE.H10MO.C |  |
Fev | 10 | C | MMGGAAATdn | HOCOMOCO v9 | 13 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 2449712 | 260298 | FEV_MOUSE | Q8QZW2 | ||||
| FLI1_MOUSE.H10MO.A |  |
Fli1 | 15 | A | vvvRCMGGAAGYdvv | ChIP-Seq | 0.8517261320114138 | 0.9301325695290349 | 3 | 399 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95554 | 14247 | FLI1_MOUSE | P26323 | |
| FOSB_MOUSE.H10MO.C |  |
Fosb | 10 | C | bRTGAKTCAK | HOCOMOCO v9 | 20 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 95575 | 14282 | FOSB_MOUSE | P13346 | ||||
| FOSL1_MOUSE.H10MO.C |  |
Fosl1 | 11 | C | nvTGACTCAbn | ChIP-Seq | 0.9555079980677658 | 0.9555079980677658 | 1 | 500 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 107179 | 14283 | FOSL1_MOUSE | P48755 | |
| FOSL2_MOUSE.H10MO.A |  |
Fosl2 | 11 | A | vvvTGAGTCAb | HOCOMOCO v9 | 2072 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 102858 | 14284 | FOSL2_MOUSE | P47930 | ||||
| FOS_MOUSE.H10MO.A |  |
Fos | 12 | A | bbvTSATTSRbb | HOCOMOCO v9 | 2087 | Fos-related factors{1.1.2} | Fos factors{1.1.2.1} | 95574 | 14281 | FOS_MOUSE | P01101 | ||||
| FOXA1_MOUSE.H10MO.B |  |
Foxa1 | 11 | B | TGTTTRCWYWb | HOCOMOCO v9 | 967 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347472 | 15375 | FOXA1_MOUSE | P35582 | ||||
| FOXA2_MOUSE.H10MO.B |  |
Foxa2 | 12 | B | hTRTTTACWYWd | ChIP-Seq | 0.8456285418078173 | 0.9159253471105573 | 2 | 502 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347476 | 15376 | FOXA2_MOUSE | P35583 | |
| FOXA3_MOUSE.H10MO.C |  |
Foxa3 | 13 | C | hTGTTTGCYYWbh | HOCOMOCO v9 | 23 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347477 | 15377 | FOXA3_MOUSE | P35584 | ||||
| FOXC1_MOUSE.H10MO.C |  |
Foxc1 | 15 | C | bnhTGTTTACWTAvS | HOCOMOCO v9 | 26 | Forkhead box (FOX) factors{3.3.1} | FOXC{3.3.1.3} | 1347466 | 17300 | FOXC1_MOUSE | Q61572 | ||||
| FOXC2_MOUSE.H10MO.D |  |
Foxc2 | 15 | D | KhTTGTTdTGYCAGd | HOCOMOCO v9 | 5 | Forkhead box (FOX) factors{3.3.1} | FOXC{3.3.1.3} | 1347481 | 14234 | FOXC2_MOUSE | Q61850 | ||||
| FOXD1_MOUSE.H10MO.D |  |
Foxd1 | 15 | D | nhhTSTTTAChWhvR | HOCOMOCO v9 | 22 | 1347463 | 15229 | FOXD1_MOUSE | Q61345 | ||||||
| FOXD3_MOUSE.H10MO.D |  |
Foxd3 | 9 | D | TGTTTRTTT | HOCOMOCO v9 | 75 | Forkhead box (FOX) factors{3.3.1} | FOXD{3.3.1.4} | 1347473 | 15221 | FOXD3_MOUSE | Q61060 | ||||
| FOXF1_MOUSE.H10MO.D |  |
Foxf1 | 11 | D | nTGTTTATbWW | HOCOMOCO v9 | 8 | Forkhead box (FOX) factors{3.3.1} | FOXF{3.3.1.6} | 1347470 | 15227 | FOXF1_MOUSE | Q61080 | ||||
| FOXF2_MOUSE.H10MO.D |  |
Foxf2 | 11 | D | TGTTTAChTWd | HOCOMOCO v9 | 26 | Forkhead box (FOX) factors{3.3.1} | FOXF{3.3.1.6} | 1347479 | 14238 | FOXF2_MOUSE | O54743 | ||||
| FOXI1_MOUSE.H10MO.B |  |
Foxi1 | 12 | B | hWbTGATTGGYb | HOCOMOCO v9 | 210 | Forkhead box (FOX) factors{3.3.1} | FOXI{3.3.1.9} | 1096329 | 14233 | FOXI1_MOUSE | Q922I5 | ||||
| FOXJ2_MOUSE.H10MO.C |  |
Foxj2 | 10 | C | TRTTTATYTd | HOCOMOCO v9 | 41 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 1926805 | 60611 | FOXJ2_MOUSE | Q9ES18 | ||||
| FOXJ3_MOUSE.H10MO.A |  |
Foxj3 | 13 | A | dTGTTTATKKTTd | HOCOMOCO v9 | 550 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 2443432 | 230700 | FOXJ3_MOUSE | Q8BUR3 | ||||
| FOXJ3_MOUSE.H10MO.S |  |
Foxj3 | 13 | S | WKKTTWWTGTTTW | HOCOMOCO v9 | 499 | Forkhead box (FOX) factors{3.3.1} | FOXJ{3.3.1.10} | 2443432 | 230700 | FOXJ3_MOUSE | Q8BUR3 | Secondary motif. | |||
| FOXM1_MOUSE.H10MO.D |  |
Foxm1 | 13 | D | bbWRTKTGWTThY | HOCOMOCO v9 | 13 | Forkhead box (FOX) factors{3.3.1} | FOXM{3.3.1.13} | 1347487 | FOXM1_MOUSE | O08696 | |||||
| FOXO1_MOUSE.H10MO.C |  |
Foxo1 | 16 | C | ddbYTGTTTWbdhndd | HOCOMOCO v9 | 58 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 1890077 | 56458 | FOXO1_MOUSE | Q9R1E0 | ||||
| FOXO3_MOUSE.H10MO.B |  |
Foxo3 | 13 | B | dRYbTGTTTWYhd | HOCOMOCO v9 | 63 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 1890081 | 56484 | FOXO3_MOUSE | Q9WVH4 | ||||
| FOXO4_MOUSE.H10MO.C |  |
Foxo4 | 9 | C | bTGTTTAbK | HOCOMOCO v9 | 28 | Forkhead box (FOX) factors{3.3.1} | FOXO{3.3.1.15} | 1891915 | 54601 | FOXO4_MOUSE | Q9WVH3 | ||||
| FOXP2_MOUSE.H10MO.A |  |
Foxp2 | 9 | A | nTGTTTMYn | HOCOMOCO v9 | 2075 | Forkhead box (FOX) factors{3.3.1} | FOXP{3.3.1.16} | 2148705 | 114142 | FOXP2_MOUSE | P58463 | ||||
| FOXP3_MOUSE.H10MO.D |  |
Foxp3 | 9 | D | WATKTGWTT | HOCOMOCO v9 | 9 | Forkhead box (FOX) factors{3.3.1} | FOXP{3.3.1.16} | 1891436 | 20371 | FOXP3_MOUSE | Q99JB6 | ||||
| FOXQ1_MOUSE.H10MO.C |  |
Foxq1 | 12 | C | nRTTGTTTATKT | HOCOMOCO v9 | 13 | Forkhead box (FOX) factors{3.3.1} | FOXQ{3.3.1.17} | 1298228 | 15220 | FOXQ1_MOUSE | O70220 | ||||
| FUBP1_MOUSE.H10MO.D |  |
Fubp1 | 12 | D | TYGTnhKTbKKK | HOCOMOCO v9 | 102 | 1196294 | 51886 | FUBP1_MOUSE | Q91WJ8 | ||||||
| GABP1_MOUSE.H10MO.C |  |
Gabpb1 | 10 | C | SCGGAAGbdv | HOCOMOCO v9 | 36 | 95611 | 14391 | GABP1_MOUSE | Q00420 | Model is applicable to several TFs or complex subunits: GABP1_MOUSE, GABP2_MOUSE. | |||||
| GABPA_MOUSE.H10MO.A |  |
Gabpa | 13 | A | vvvCCGGAAGYSv | HOCOMOCO v9 | 2926 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95610 | 14390 | GABPA_MOUSE | Q00422 | ||||
| GATA1_MOUSE.H10MO.A |  |
Gata1 | 11 | A | nvAGATAASvn | ChIP-Seq | 0.8677528789590304 | 0.9101870554711257 | 13 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95661 | 14460 | GATA1_MOUSE | P17679 | |
| GATA2_MOUSE.H10MO.A |  |
Gata2 | 11 | A | vvWGATAASvn | ChIP-Seq | 0.8357221523117732 | 0.8853869410431214 | 5 | 500 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95662 | 14461 | GATA2_MOUSE | O09100 | |
| GATA3_MOUSE.H10MO.C |  |
Gata3 | 8 | C | vWGATARv | HOCOMOCO v9 | 0.7238850047195652 | 0.7495760198196578 | 2 | 89 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95663 | 14462 | GATA3_MOUSE | P23772 | |
| GATA4_MOUSE.H10MO.B |  |
Gata4 | 9 | B | dvWGATARv | HOCOMOCO v9 | 107 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95664 | 14463 | GATA4_MOUSE | Q08369 | ||||
| GATA5_MOUSE.H10MO.D |  |
Gata5 | 17 | D | WvMnWGATAWbnhdRhd | HOCOMOCO v9 | 13 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 109497 | 14464 | GATA5_MOUSE | P97489 | ||||
| GATA6_MOUSE.H10MO.B |  |
Gata6 | 7 | B | nhGATAR | HOCOMOCO v9 | 115 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 107516 | 14465 | GATA6_MOUSE | Q61169 | ||||
| GCM1_MOUSE.H10MO.D |  |
Gcm1 | 14 | D | hWnATGMKGGYhbb | HOCOMOCO v9 | 12 | GCM factors{7.2.1} | GCMa (GCM1){7.2.1.0.1} | 108045 | 14531 | GCM1_MOUSE | P70348 | ||||
| GCR_MOUSE.H10MO.C |  |
Nr3c1 | 18 | C | vRGRASAdnvhRdnSndd | HOCOMOCO v9 | 0.7467282195273168 | 0.757625051561599 | 2 | 4198 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 95824 | GCR_MOUSE | P06537 | ||
| GCR_MOUSE.H10MO.S |  |
Nr3c1 | 7 | S | RGRACAd | HOCOMOCO v9 | 0.6591818067108669 | 0.6689139877791731 | 2 | 4629 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 95824 | GCR_MOUSE | P06537 | Secondary motif. | |
| GFI1B_MOUSE.H10MO.C |  |
Gfi1b | 11 | C | RCWSWGRTTKv | ChIP-Seq | 0.8213722759493574 | 0.8213722759493574 | 1 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 1276578 | 14582 | GFI1B_MOUSE | O70237 | |
| GFI1_MOUSE.H10MO.C |  |
Gfi1 | 10 | C | RShSWGATTb | HOCOMOCO v9 | 152 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 103170 | 14581 | GFI1_MOUSE | P70338 | ||||
| GLI1_MOUSE.H10MO.C |  |
Gli1 | 11 | C | bTGGGdGGTCb | HOCOMOCO v9 | 31 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 95727 | 14632 | GLI1_MOUSE | P47806 | ||||
| GLI2_MOUSE.H10MO.B |  |
Gli2 | 11 | B | STGGGTGGTCb | HOCOMOCO v9 | 970 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 95728 | 14633 | GLI2_MOUSE | Q0VGT2 | ||||
| GLI3_MOUSE.H10MO.B |  |
Gli3 | 11 | B | nTGGGTGGTYb | HOCOMOCO v9 | 15 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 95729 | 14634 | GLI3_MOUSE | Q61602 | ||||
| GLIS3_MOUSE.H10MO.D |  |
Glis3 | 10 | D | SYGGGGGGTM | HOCOMOCO v9 | 38 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 2444289 | 226075 | GLIS3_MOUSE | Q6XP49 | ||||
| HAND1_MOUSE.H10MO.D |  |
Hand1 | 12 | D | dnRTCTGGhWdh | HOCOMOCO v9 | 26 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 103577 | 15110 | HAND1_MOUSE | Q64279 | ||||
| HBP1_MOUSE.H10MO.D |  |
Hbp1 | 9 | D | RnYSATTGA | HOCOMOCO v9 | 5 | SOX-related factors{4.1.1} | Further Sox-related factors{4.1.1.9} | 894659 | 73389 | HBP1_MOUSE | Q8R316 | ||||
| HEN1_MOUSE.H10MO.C |  |
Nhlh1 | 20 | C | bKGGnMbCAGCTGCGKCYhh | HOCOMOCO v9 | 55 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 98481 | 18071 | HEN1_MOUSE | Q02576 | ||||
| HES1_MOUSE.H10MO.D |  |
Hes1 | 13 | D | vMSvCAMKvGMhv | HOCOMOCO v9 | 28 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 104853 | 15205 | HES1_MOUSE | P35428 | ||||
| HESX1_MOUSE.H10MO.D |  |
Hesx1 | 16 | D | nKCYGRCWCnTRRnhY | HOCOMOCO v9 | 8 | Paired-related HD factors{3.1.3} | HESX{3.1.3.10} | 96071 | 15209 | HESX1_MOUSE | Q61658 | ||||
| HEY2_MOUSE.H10MO.D |  |
Hey2 | 17 | D | GKKSGCWCGTGSCnYKv | HOCOMOCO v9 | 8 | Hairy-related factors{1.2.4} | Hairy-like factors{1.2.4.1} | 1341884 | 15214 | HEY2_MOUSE | Q9QUS4 | ||||
| HIC1_MOUSE.H10MO.C |  |
Hic1 | 9 | C | dKGKTGCCM | HOCOMOCO v9 | 40 | Factors with multiple dispersed zinc fingers{2.3.4} | Hypermethylated in Cancer proteins{2.3.4.17} | 1338010 | 15248 | HIC1_MOUSE | Q9R1Y5 | ||||
| HIF1A_MOUSE.H10MO.A |  |
Hif1a | 12 | A | vbvbACGTGMvb | HOCOMOCO v9 | 501 | PAS domain factors{1.2.5} | Ahr-like factors{1.2.5.1} | 106918 | 15251 | HIF1A_MOUSE | Q61221 | ||||
| HINFP_MOUSE.H10MO.C |  |
Hinfp | 16 | C | dhSnnvdCGGACGTWv | HOCOMOCO v9 | 42 | Factors with multiple dispersed zinc fingers{2.3.4} | HINFP-like factors{2.3.4.21} | 2429620 | 102423 | HINFP_MOUSE | Q8K1K9 | ||||
| HLF_MOUSE.H10MO.C |  |
Hlf | 13 | C | nRTKRYGTAAYvn | HOCOMOCO v9 | 35 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 96108 | 217082 | HLF_MOUSE | Q8BW74 | ||||
| HLTF_MOUSE.H10MO.D |  |
Hltf | 12 | D | nAnGKCWGCAAM | HOCOMOCO v9 | 10 | 1196437 | 20585 | HLTF_MOUSE | Q6PCN7 | ||||||
| HMGA1_MOUSE.H10MO.D |  |
Hmga1 | 7 | D | nWATTWd | HOCOMOCO v9 | 97 | HMGA factors{8.2.1} | HMGA1 (HMGI(Y)){8.2.1.0.1} | 96160 | 111241; 15361 | HMGA1_MOUSE | P17095 | ||||
| HMGA2_MOUSE.H10MO.D |  |
Hmga2 | 15 | D | AATAWhSSSSAATAT | HOCOMOCO v9 | 75 | HMGA factors{8.2.1} | HMGA2 (HMGI-C){8.2.1.0.2} | 101761 | 15364 | HMGA2_MOUSE | P52927 | ||||
| HNF1A_MOUSE.H10MO.A |  |
Hnf1a | 15 | A | RGTTAWTvWTYdhYv | HOCOMOCO v9 | 175 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 98504 | 21405 | HNF1A_MOUSE | P22361 | ||||
| HNF1B_MOUSE.H10MO.B |  |
Hnf1b | 14 | B | dRTTAATRdTTdAY | HOCOMOCO v9 | 58 | POU domain factors{3.1.10} | HNF1-like factors{3.1.10.7} | 98505 | 21410 | HNF1B_MOUSE | P27889 | ||||
| HNF4A_MOUSE.H10MO.A |  |
Hnf4a | 13 | A | dGdbCAAAGKbCR | HOCOMOCO v9 | 1867 | RXR-related receptors (NR2){2.1.3} | HNF-4 (NR2A){2.1.3.2} | 109128 | 15378 | HNF4A_MOUSE | P49698 | ||||
| HNF4G_MOUSE.H10MO.C |  |
Hnf4g | 12 | C | SGSCAMAGYMCA | HOCOMOCO v9 | 7 | RXR-related receptors (NR2){2.1.3} | HNF-4 (NR2A){2.1.3.2} | 1353604 | 30942 | HNF4G_MOUSE | Q9WUU6 | ||||
| HNF6_MOUSE.H10MO.C |  |
Onecut1 | 14 | C | dWYATTGATTWhdh | HOCOMOCO v9 | 24 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 1196423 | 15379 | HNF6_MOUSE | O08755 | ||||
| HSF1_MOUSE.H10MO.A |  |
Hsf1 | 14 | A | vGAAnnWWSbRGMR | HOCOMOCO v9 | 2083 | HSF factors{3.4.1} | HSF1 (HSTF1){3.4.1.0.1} | 96238 | 15499 | HSF1_MOUSE | P38532 | ||||
| HSF2_MOUSE.H10MO.A |  |
Hsf2 | 14 | A | vSRWbvWKSKvGRh | HOCOMOCO v9 | 1043 | HSF factors{3.4.1} | HSF2 (HSTF2){3.4.1.0.2} | 96239 | 15500 | HSF2_MOUSE | P38533 | ||||
| HTF4_MOUSE.H10MO.C |  |
Tcf12 | 11 | C | vvvRRCAGCTG | ChIP-Seq | 0.9279493092210841 | 0.927949309221084 | 1 | 501 | E2A-related factors{1.2.1} | HTF-4 (TCF-12, HEB){1.2.1.0.3} | 101877 | 21406 | HTF4_MOUSE | Q61286 | |
| HXA10_MOUSE.H10MO.C |  |
Hoxa10 | 12 | C | dnYnRTTTATGh | HOCOMOCO v9 | 10 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96171 | 15395 | HXA10_MOUSE | P31310 | ||||
| HXA13_MOUSE.H10MO.C |  |
Hoxa13 | 11 | C | vdTWTTATKGS | HOCOMOCO v9 | 18 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96173 | 15398 | HXA13_MOUSE | Q62424 | ||||
| HXA1_MOUSE.H10MO.C |  |
Hoxa1 | 10 | C | nTGATGGATG | HOCOMOCO v9 | 8 | HOX-related factors{3.1.1} | HOX1{3.1.1.1} | 96170 | HXA1_MOUSE | P09022 | |||||
| HXA5_MOUSE.H10MO.D |  |
Hoxa5 | 10 | D | YTvATTARYR | HOCOMOCO v9 | 54 | HOX-related factors{3.1.1} | HOX5{3.1.1.5} | 96177 | 15402 | HXA5_MOUSE | P09021 | ||||
| HXA7_MOUSE.H10MO.D | 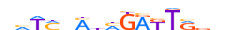 |
Hoxa7 | 15 | D | vhTMnMnMGAKTRvv | HOCOMOCO v9 | 21 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 96179 | 15404 | HXA7_MOUSE | P02830 | ||||
| HXA9_MOUSE.H10MO.D |  |
Hoxa9 | 14 | D | TGACASKTTWAYRR | HOCOMOCO v9 | 29 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96180 | 15405 | HXA9_MOUSE | P09631 | ||||
| HXB1_MOUSE.H10MO.D |  |
Hoxb1 | 10 | D | WGATGGATGG | HOCOMOCO v9 | 15 | HOX-related factors{3.1.1} | HOX1{3.1.1.1} | 96182 | 15407 | HXB1_MOUSE | P17919 | ||||
| HXB6_MOUSE.H10MO.D |  |
Hoxb6 | 13 | D | dWTGATTKATGCn | HOCOMOCO v9 | 7 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 96187 | 15414 | HXB6_MOUSE | P09023 | ||||
| HXB7_MOUSE.H10MO.C |  |
Hoxb7 | 10 | C | WTGATTRATK | HOCOMOCO v9 | 28 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 96188 | 15415 | HXB7_MOUSE | P09024 | ||||
| HXB8_MOUSE.H10MO.C |  |
Hoxb8 | 11 | C | TTGATTRATKv | HOCOMOCO v9 | 26 | HOX-related factors{3.1.1} | HOX8{3.1.1.7} | 96189 | 15416 | HXB8_MOUSE | P09632 | ||||
| HXC6_MOUSE.H10MO.D |  |
Hoxc6 | 15 | D | WTGATTTATTWCTTT | HOCOMOCO v9 | 7 | HOX-related factors{3.1.1} | HOX6-7{3.1.1.6} | 96197 | 15425 | HXC6_MOUSE | P10629 | ||||
| HXC8_MOUSE.H10MO.D |  |
Hoxc8 | 14 | D | nTTSATTRAnKSnn | HOCOMOCO v9 | 9 | HOX-related factors{3.1.1} | HOX8{3.1.1.7} | 96198 | 15426 | HXC8_MOUSE | P09025 | ||||
| HXD10_MOUSE.H10MO.D |  |
Hoxd10 | 10 | D | TGnTYTWRTK | HOCOMOCO v9 | 7 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96202 | 15430 | HXD10_MOUSE | P28359 | ||||
| HXD13_MOUSE.H10MO.D |  |
Hoxd13 | 11 | D | WTTAYKRKRGM | HOCOMOCO v9 | 13 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96205 | 15433 | HXD13_MOUSE | P70217 | ||||
| HXD4_MOUSE.H10MO.D |  |
Hoxd4 | 8 | D | KTAATTnW | HOCOMOCO v9 | 8 | HOX-related factors{3.1.1} | HOX4{3.1.1.4} | 96208 | 15436 | HXD4_MOUSE | P10628 | ||||
| HXD9_MOUSE.H10MO.D |  |
Hoxd9 | 10 | D | AnWTTnATnn | HOCOMOCO v9 | 6 | HOX-related factors{3.1.1} | HOX9-13{3.1.1.8} | 96210 | 15438 | HXD9_MOUSE | P28357 | ||||
| IKZF1_MOUSE.H10MO.C |  |
Ikzf1 | 8 | C | bTGGGARd | HOCOMOCO v9 | 61 | Factors with multiple dispersed zinc fingers{2.3.4} | Ikaros{2.3.4.4} | 1342540 | IKZF1_MOUSE | Q03267 | |||||
| INSM1_MOUSE.H10MO.C |  |
Insm1 | 12 | C | KGYMAGGGGGCA | HOCOMOCO v9 | 24 | Factors with multiple dispersed zinc fingers{2.3.4} | Insulinoma-associated proteins{2.3.4.16} | 1859980 | 53626 | INSM1_MOUSE | Q63ZV0 | ||||
| IRF1_MOUSE.H10MO.C |  |
Irf1 | 12 | C | RAAAvYGAAAbh | HOCOMOCO v9 | 135 | Interferon-regulatory factors{3.5.3} | IRF-1{3.5.3.0.1} | 96590 | 16362 | IRF1_MOUSE | P15314 | ||||
| IRF2_MOUSE.H10MO.C |  |
Irf2 | 14 | C | bRAAARYGAAASbv | HOCOMOCO v9 | 84 | Interferon-regulatory factors{3.5.3} | IRF-2{3.5.3.0.2} | 96591 | 16363 | IRF2_MOUSE | P23906 | ||||
| IRF3_MOUSE.H10MO.C |  |
Irf3 | 17 | C | RRAAASbGAAAvbdvdh | HOCOMOCO v9 | 46 | Interferon-regulatory factors{3.5.3} | IRF-3{3.5.3.0.3} | 1859179 | 54131 | IRF3_MOUSE | P70671 | ||||
| IRF4_MOUSE.H10MO.C |  |
Irf4 | 16 | C | vWRRvRGRAAMhRAAA | HOCOMOCO v9 | 2115 | Interferon-regulatory factors{3.5.3} | IRF-4 (LSIRF, NF-EM5, MUM1, Pip){3.5.3.0.4} | 1096873 | 16364 | IRF4_MOUSE | Q64287 | ||||
| IRF5_MOUSE.H10MO.D |  |
Irf5 | 20 | D | KAAAGGAAAGCnAAAAGTGM | HOCOMOCO v9 | 5 | Interferon-regulatory factors{3.5.3} | IRF-5{3.5.3.0.5} | 1350924 | 27056 | IRF5_MOUSE | P56477 | ||||
| IRF7_MOUSE.H10MO.C | 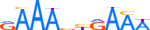 |
Irf7 | 10 | C | RAAAbYRAAW | HOCOMOCO v9 | 40 | Interferon-regulatory factors{3.5.3} | IRF-7{3.5.3.0.7} | 1859212 | 54123 | IRF7_MOUSE | P70434 | ||||
| IRF8_MOUSE.H10MO.D |  |
Irf8 | 15 | D | vvRAAAvYGAAAvYv | HOCOMOCO v9 | 37 | Interferon-regulatory factors{3.5.3} | IRF-8 (ICSBP1){3.5.3.0.8} | 96395 | 15900 | IRF8_MOUSE | P23611 | ||||
| IRF9_MOUSE.H10MO.C |  |
Irf9 | 12 | C | GAAAbhGAAACT | HOCOMOCO v9 | 5 | Interferon-regulatory factors{3.5.3} | IRF-9 (ISGF-3gamma){3.5.3.0.9} | 107587 | 16391 | IRF9_MOUSE | Q61179 | ||||
| ISL1_MOUSE.H10MO.D |  |
Isl1 | 7 | D | SYTAATG | HOCOMOCO v9 | 13 | HD-LIM factors{3.1.5} | ISL{3.1.5.1} | 101791 | 16392 | ISL1_MOUSE | P61372 | ||||
| ITF2_MOUSE.H10MO.B |  |
Tcf4 | 9 | B | vCAGGTGYd | HOCOMOCO v9 | 553 | E2A-related factors{1.2.1} | SEF2 (E2-2, TCF-4, ITF-2, MITF){1.2.1.0.2} | 98506 | 21413 | ITF2_MOUSE | Q60722 | ||||
| JUNB_MOUSE.H10MO.B |  |
Junb | 10 | B | dvTGAGTCAb | HOCOMOCO v9 | 58 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96647 | 16477 | JUNB_MOUSE | P09450 | ||||
| JUND_MOUSE.H10MO.A |  |
Jund | 11 | A | vvTGAGTCAYh | HOCOMOCO v9 | 0.7677977610264641 | 0.7677977610264641 | 1 | 4129 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96648 | 16478 | JUND_MOUSE | P15066 | |
| JUN_MOUSE.H10MO.A |  |
Jun | 9 | A | vTGAGTCAb | HOCOMOCO v9 | 0.7257210624644428 | 0.7257210624644428 | 1 | 2361 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96646 | 16476 | JUN_MOUSE | P05627 | |
| KAISO_MOUSE.H10MO.B |  |
Zbtb33 | 12 | B | hTvGCARGAddn | HOCOMOCO v9 | 2038 | Other factors with up to three adjacent zinc fingers{2.3.2} | Factors with 2-3 adjacent zinc fingersand a BTB/POZ domain{2.3.2.1} | 1927290 | 56805 | KAISO_MOUSE | Q8BN78 | ||||
| KLF15_MOUSE.H10MO.D |  |
Klf15 | 17 | D | RShKGGGAGdKKGSGSS | HOCOMOCO v9 | 11 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1929988 | 66277 | KLF15_MOUSE | Q9EPW2 | ||||
| KLF1_MOUSE.H10MO.C |  |
Klf1 | 11 | C | bWGGGTGnGGb | HOCOMOCO v9 | 11 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342771 | KLF1_MOUSE | P46099 | |||||
| KLF3_MOUSE.H10MO.D |  |
Klf3 | 15 | D | WRChWGGGTGTSGnW | HOCOMOCO v9 | 8 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342773 | 16599 | KLF3_MOUSE | Q60980 | ||||
| KLF4_MOUSE.H10MO.C |  |
Klf4 | 12 | C | WGGGYGKGGYbb | ChIP-Seq | 0.9234290797584314 | 0.9234290797584314 | 1 | 501 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342287 | 16600 | KLF4_MOUSE | Q60793 | |
| KLF6_MOUSE.H10MO.D |  |
Klf6 | 8 | D | SGGGSCGG | HOCOMOCO v9 | 11 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1346318 | KLF6_MOUSE | O08584 | |||||
| KLF8_MOUSE.H10MO.C |  |
Klf8 | 9 | C | CAGGGGGTG | HOCOMOCO v9 | 5 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 2442430 | 245671 | KLF8_MOUSE | Q8BLM0 | ||||
| LEF1_MOUSE.H10MO.C |  |
Lef1 | 7 | C | CTTTGWW | HOCOMOCO v9 | 67 | TCF-7-related factors{4.1.3} | LEF-1 (TCF-1alpha) [1]{4.1.3.0.4} | 96770 | 16842 | LEF1_MOUSE | P27782 | ||||
| LHX2_MOUSE.H10MO.D |  |
Lhx2 | 13 | D | hdnWTKTAATWAS | HOCOMOCO v9 | 7 | HD-LIM factors{3.1.5} | Lhx-2-like factors{3.1.5.3} | 96785 | 16870 | LHX2_MOUSE | Q9Z0S2 | ||||
| LHX3_MOUSE.H10MO.C |  |
Lhx3 | 13 | C | ddAWTTAATTAdh | HOCOMOCO v9 | 72 | HD-LIM factors{3.1.5} | Lhx-3-like factors{3.1.5.4} | 102673 | 16871 | LHX3_MOUSE | P50481 | ||||
| MAFA_MOUSE.H10MO.D |  |
Mafa | 21 | D | bKGCTGWMShhSYWvYYWYYv | HOCOMOCO v9 | 27 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 2673307 | 378435 | MAFA_MOUSE | Q8CF90 | ||||
| MAFB_MOUSE.H10MO.D |  |
Mafb | 9 | D | WGMTGASdb | HOCOMOCO v9 | 48 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 104555 | 16658 | MAFB_MOUSE | P54841 | ||||
| MAFG_MOUSE.H10MO.D |  |
Mafg | 7 | D | hGTCATd | HOCOMOCO v9 | 66 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 96911 | 17134 | MAFG_MOUSE | O54790 | ||||
| MAFK_MOUSE.H10MO.A |  |
Mafk | 18 | A | ddWWnTGCTGASTMAKCW | ChIP-Seq | 0.9563854263905045 | 0.9608432559648592 | 2 | 441 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 99951 | 17135 | MAFK_MOUSE | Q61827 | |
| MAFK_MOUSE.H10MO.S |  |
Mafk | 10 | S | YGCTGAGTCA | HOCOMOCO v9 | 0.8977825346888878 | 0.9049094684454113 | 2 | 17 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 99951 | 17135 | MAFK_MOUSE | Q61827 | Secondary motif. |
| MAF_MOUSE.H10MO.B |  |
Maf | 8 | B | dYGCTGAS | HOCOMOCO v9 | 62 | Maf-related factors{1.1.3} | Large Maf factors{1.1.3.1} | 96909 | 17132 | MAF_MOUSE | P54843 | ||||
| MAX_MOUSE.H10MO.A |  |
Max | 11 | A | vvvCACGTGvY | ChIP-Seq | 0.8256301075981137 | 0.9014373775933924 | 4 | 442 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 96921 | MAX_MOUSE | P28574 | ||
| MAZ_MOUSE.H10MO.D |  |
Maz | 17 | D | dSRGdRRGGRGGRRRGS | HOCOMOCO v9 | 31 | Factors with multiple dispersed zinc fingers{2.3.4} | MAZ-like factors{2.3.4.8} | 1338823 | MAZ_MOUSE | P56671 | |||||
| MBD2_MOUSE.H10MO.B |  |
Mbd2 | 11 | B | nSGGCCGGMKv | HOCOMOCO v9 | 113 | 1333813 | 17191 | MBD2_MOUSE | Q9Z2E1 | ||||||
| MCR_MOUSE.H10MO.D |  |
Nr3c2 | 17 | D | hAGWMCARSTKGTdGKR | HOCOMOCO v9 | 8 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 99459 | MCR_MOUSE | Q8VII8 | |||||
| MECP2_MOUSE.H10MO.C |  |
Mecp2 | 7 | C | SCCGGAG | HOCOMOCO v9 | 122 | 99918 | 17257 | MECP2_MOUSE | Q9Z2D6 | ||||||
| MEF2A_MOUSE.H10MO.B |  |
Mef2a | 14 | B | dddYTATTTATAGh | HOCOMOCO v9 | 143 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99532 | 17258 | MEF2A_MOUSE | Q60929 | ||||
| MEF2C_MOUSE.H10MO.B |  |
Mef2c | 13 | B | ddCTATAAATAGh | HOCOMOCO v9 | 563 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99458 | 17260 | MEF2C_MOUSE | Q8CFN5 | ||||
| MEF2D_MOUSE.H10MO.C |  |
Mef2d | 12 | C | KCTATTTTTAKv | HOCOMOCO v9 | 11 | Regulators of differentiation{5.1.1} | MEF-2{5.1.1.1} | 99533 | 17261 | MEF2D_MOUSE | Q63943 | ||||
| MEIS1_MOUSE.H10MO.C |  |
Meis1 | 13 | C | TGACAGbTKWMhd | HOCOMOCO v9 | 40 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 104717 | 17268 | MEIS1_MOUSE | Q60954 | ||||
| MEIS2_MOUSE.H10MO.B |  |
Meis2 | 13 | B | hTGACAGbTKTbv | HOCOMOCO v9 | 413 | TALE-type homeo domain factors{3.1.4} | MEIS{3.1.4.2} | 108564 | 17536 | MEIS2_MOUSE | P97367 | ||||
| MITF_MOUSE.H10MO.C |  |
Mitf | 10 | C | vhCAYGTGvb | HOCOMOCO v9 | 69 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 104554 | 17342 | MITF_MOUSE | Q08874 | ||||
| MLXPL_MOUSE.H10MO.D |  |
Mlxipl | 19 | D | SCAYGKGRMMMYnbSGTGb | HOCOMOCO v9 | 11 | bHLH-ZIP factors{1.2.6} | Mondo-like factors{1.2.6.6} | 1927999 | 58805 | MLXPL_MOUSE | Q99MZ3 | ||||
| MSX2_MOUSE.H10MO.D |  |
Msx2 | 7 | D | TAATTnb | HOCOMOCO v9 | 12 | NK-related factors{3.1.2} | MSX{3.1.2.11} | 97169 | 17702 | MSX2_MOUSE | Q03358 | ||||
| MSX3_MOUSE.H10MO.D |  |
Msx3 | 17 | D | TAATTGbWWdWTAATTd | HT-SELEX | 2675 | 106587 | 17703 | MSX3_MOUSE | P70354 | ||||||
| MTF1_MOUSE.H10MO.C |  |
Mtf1 | 17 | C | vGKRMCGKGTGCAdMdb | HOCOMOCO v9 | 33 | More than 3 adjacent zinc finger factors{2.3.3} | MTF1-like factors{2.3.3.24} | 101786 | 17764 | MTF1_MOUSE | Q07243 | ||||
| MXI1_MOUSE.H10MO.C | 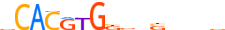 |
Mxi1 | 15 | C | vCACRYGKvnSbbbv | ChIP-Seq | 0.7623422759406301 | 0.7683614480295263 | 2 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 97245 | 17859 | MXI1_MOUSE | P50540 | |
| MYBB_MOUSE.H10MO.D |  |
Mybl2 | 10 | D | dRSAnGTTdW | HOCOMOCO v9 | 9 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 101785 | 17865 | MYBB_MOUSE | P48972 | ||||
| MYB_MOUSE.H10MO.C |  |
Myb | 11 | C | vdKRCMGTTRv | ChIP-Seq | 0.7934293636126373 | 0.7987933753601197 | 2 | 500 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 97249 | 17863 | MYB_MOUSE | P06876 | |
| MYCN_MOUSE.H10MO.B |  |
Mycn | 8 | B | MCRCGTGv | HOCOMOCO v9 | 0.6918903718611138 | 0.6918903718611138 | 1 | 553 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 97357 | 18109 | MYCN_MOUSE | P03966 | |
| MYC_MOUSE.H10MO.A |  |
Myc | 12 | A | vvdvvCACRTGK | ChIP-Seq | 0.8510897707174321 | 0.883466684827523 | 3 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 97250 | 17869 | MYC_MOUSE | P01108 | |
| MYF6_MOUSE.H10MO.C |  |
Myf6 | 7 | C | GCAGCTG | HOCOMOCO v9 | 9 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97253 | 17878 | MYF6_MOUSE | P15375 | ||||
| MYOD1_MOUSE.H10MO.C |  |
Myod1 | 15 | C | vCAGCTGYYbYhbbY | ChIP-Seq | 0.9231358015382102 | 0.9231358015382103 | 1 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97275 | 17927 | MYOD1_MOUSE | P10085 | |
| MYOG_MOUSE.H10MO.C |  |
Myog | 11 | C | vvvRRCAGCTG | ChIP-Seq | 0.936904934324688 | 0.936904934324688 | 1 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97276 | 17928 | MYOG_MOUSE | P12979 | |
| NANOG_MOUSE.H10MO.A |  |
Nanog | 17 | A | bbYWTTvWnWTKYWRAd | ChIP-Seq | 0.782272397117236 | 0.8499834054583915 | 4 | 500 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 1919200 | 71950 | NANOG_MOUSE | Q80Z64 | |
| NANOG_MOUSE.H10MO.S |  |
Nanog | 8 | S | TTTAATGb | HOCOMOCO v9 | 0.49859151249281575 | 0.5817741120218001 | 4 | 5 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 1919200 | 71950 | NANOG_MOUSE | Q80Z64 | Secondary motif. |
| NDF1_MOUSE.H10MO.C |  |
Neurod1 | 11 | C | nRRCAGMTGGb | ChIP-Seq | 0.8718431704713612 | 0.8718431704713613 | 1 | 478 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1339708 | 18012 | NDF1_MOUSE | Q60867 | |
| NF2L1_MOUSE.H10MO.C |  |
Nfe2l1 | 7 | C | nRTGAYd | HOCOMOCO v9 | 115 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 99421 | 18023 | NF2L1_MOUSE | Q61985 | ||||
| NF2L2_MOUSE.H10MO.D |  |
Nfe2l2 | 12 | D | vRTGAYWMRGCM | HOCOMOCO v9 | 92 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 108420 | 18024 | NF2L2_MOUSE | Q60795 | ||||
| NFAC1_MOUSE.H10MO.A |  |
Nfatc1 | 12 | A | hhRYGGAAWdWY | HOCOMOCO v9 | 756 | NFAT-related factors{6.1.3} | NFATc1{6.1.3.0.1} | 102469 | NFAC1_MOUSE | O88942 | |||||
| NFAC1_MOUSE.H10MO.S |  |
Nfatc1 | 13 | S | hGGAAWdTTCMWW | HOCOMOCO v9 | 558 | NFAT-related factors{6.1.3} | NFATc1{6.1.3.0.1} | 102469 | NFAC1_MOUSE | O88942 | Secondary motif. | ||||
| NFAC2_MOUSE.H10MO.B |  |
Nfatc2 | 9 | B | dGGAAAWhb | HOCOMOCO v9 | 87 | NFAT-related factors{6.1.3} | NFATc2 (NFATp, NFAT1){6.1.3.0.2} | 102463 | 18019 | NFAC2_MOUSE | Q60591 | ||||
| NFAC3_MOUSE.H10MO.B |  |
Nfatc3 | 9 | B | hGGAAAAhY | HOCOMOCO v9 | 18 | NFAT-related factors{6.1.3} | NFATc3 (NFATx, NFAT4){6.1.3.0.3} | 103296 | NFAC3_MOUSE | P97305 | |||||
| NFAC4_MOUSE.H10MO.C |  |
Nfatc4 | 10 | C | dGGAAARhdW | HOCOMOCO v9 | 19 | NFAT-related factors{6.1.3} | NFATc4 (NFAT3){6.1.3.0.4} | 1920431 | 73181 | NFAC4_MOUSE | Q8K120 | ||||
| NFAT5_MOUSE.H10MO.D |  |
Nfat5 | 15 | D | STGGAAAAnYYCMKK | HOCOMOCO v9 | 8 | NFAT-related factors{6.1.3} | NFAT5 (TonEBP){6.1.3.0.5} | 1859333 | 54446 | NFAT5_MOUSE | Q9WV30 | ||||
| NFE2_MOUSE.H10MO.A |  |
Nfe2 | 14 | A | vvvRTGASTCRGCW | HOCOMOCO v9 | 1435 | Jun-related factors{1.1.1} | NF-E2-like factors{1.1.1.2} | 97308 | 18022 | NFE2_MOUSE | Q07279 | ||||
| NFIA_MOUSE.H10MO.C |  |
Nfia | 15 | C | hTGGShnGnhKCCWd | HOCOMOCO v9 | 233 | Nuclear factor 1{7.1.2} | NF-1A (NF-IA){7.1.2.0.1} | 108056 | 18027 | NFIA_MOUSE | Q02780 | Model is applicable to several TFs or complex subunits: NFIA_MOUSE, NFIB_MOUSE, NFIC_MOUSE, NFIX_MOUSE. | |||
| NFIA_MOUSE.H10MO.S |  |
Nfia | 7 | S | hYTGGCd | HOCOMOCO v9 | 267 | Nuclear factor 1{7.1.2} | NF-1A (NF-IA){7.1.2.0.1} | 108056 | 18027 | NFIA_MOUSE | Q02780 | Model is applicable to several TFs or complex subunits: NFIA_MOUSE, NFIB_MOUSE, NFIC_MOUSE, NFIX_MOUSE. Secondary motif. | |||
| NFIL3_MOUSE.H10MO.C |  |
Nfil3 | 14 | C | vTTAYGTAAbbbvb | HOCOMOCO v9 | 51 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 109495 | 18030 | NFIL3_MOUSE | O08750 | ||||
| NFKB1_MOUSE.H10MO.B |  |
Nfkb1 | 11 | B | dGGRAAKYCCC | HOCOMOCO v9 | 185 | NF-kappaB-related factors{6.1.1} | NF-kappaB p50 subunit-like factors{6.1.1.1} | 97312 | 18033 | NFKB1_MOUSE | P25799 | ||||
| NFKB2_MOUSE.H10MO.D | 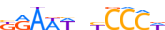 |
Nfkb2 | 11 | D | bGAWWbYCCCW | HOCOMOCO v9 | 11 | NF-kappaB-related factors{6.1.1} | NF-kappaB p50 subunit-like factors{6.1.1.1} | 1099800 | 18034 | NFKB2_MOUSE | Q9WTK5 | ||||
| NFYA_MOUSE.H10MO.D |  |
Nfya | 21 | D | YKSYSATTKKYYvdbdbbbbY | ChIP-Seq | 0.7330768639727392 | 0.7330768639727391 | 1 | 482 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YA (CP1A, CBF-B){4.2.1.0.1} | 97316 | 18044 | NFYA_MOUSE | P23708 | |
| NFYA_MOUSE.H10MO.S |  |
Nfya | 13 | S | hbSYSATTGGYYv | HOCOMOCO v9 | 0.6207241796019873 | 0.6207241796019873 | 1 | 1318 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YA (CP1A, CBF-B){4.2.1.0.1} | 97316 | 18044 | NFYA_MOUSE | P23708 | Secondary motif. |
| NFYB_MOUSE.H10MO.A |  |
Nfyb | 13 | A | bRRCCAATSRSvd | HOCOMOCO v9 | 1163 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YB (CP1B, CBF-A){4.2.1.0.2} | 97317 | 18045 | NFYB_MOUSE | P63139 | ||||
| NFYC_MOUSE.H10MO.B |  |
Nfyc | 14 | B | hRRCCAATSRSnvv | HOCOMOCO v9 | 26 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YC{4.2.1.0.3} | 107901 | 18046 | NFYC_MOUSE | P70353 | ||||
| NKX21_MOUSE.H10MO.D | 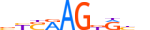 |
Nkx2-1 | 10 | D | bYSRAGdRbh | HOCOMOCO v9 | 85 | NK-related factors{3.1.2} | NK-2.1{3.1.2.14} | 108067 | 21869 | NKX21_MOUSE | P50220 | ||||
| NKX22_MOUSE.H10MO.D |  |
Nkx2-2 | 12 | D | bWYAAGTRbTTd | HOCOMOCO v9 | 25 | NK-related factors{3.1.2} | NK-2.2{3.1.2.15} | 97347 | 18088 | NKX22_MOUSE | P42586 | ||||
| NKX25_MOUSE.H10MO.C |  |
Nkx2-5 | 7 | C | KbAAGTG | HOCOMOCO v9 | 60 | NK-related factors{3.1.2} | NK-4{3.1.2.17} | 97350 | 18091 | NKX25_MOUSE | P42582 | ||||
| NKX28_MOUSE.H10MO.C |  |
Nkx2-8 | 9 | C | bTCAAGGAb | HOCOMOCO v9 | 6 | NK-related factors{3.1.2} | NK-2.2{3.1.2.15} | 1270158 | 18094 | NKX28_MOUSE | O70584 | ||||
| NKX31_MOUSE.H10MO.C |  |
Nkx3-1 | 13 | C | WWTAAGTAWWhdh | HOCOMOCO v9 | 43 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 97352 | 18095 | NKX31_MOUSE | P97436 | ||||
| NKX32_MOUSE.H10MO.C |  |
Nkx3-2 | 11 | C | nRYWAAGTGSR | HOCOMOCO v9 | 19 | NK-related factors{3.1.2} | NK-3{3.1.2.16} | 108015 | 12020 | NKX32_MOUSE | P97503 | ||||
| NOBOX_MOUSE.H10MO.C |  |
Nobox | 9 | C | hTAATTGvb | HOCOMOCO v9 | 66 | 108011 | 18291 | NOBOX_MOUSE | Q8VIH1 | ||||||
| NR0B1_MOUSE.H10MO.D |  |
Nr0b1 | 10 | D | SCGKGGRASA | HOCOMOCO v9 | 9 | DAX-related receptors (NR0){2.1.7} | DAX1 (NR0B1){2.1.7.0.1} | 1352460 | 11614 | NR0B1_MOUSE | Q61066 | ||||
| NR1D1_MOUSE.H10MO.D |  |
Nr1d1 | 19 | D | vdRdvvvnAGGbCAvdddv | ChIP-Seq | 0.7223133247224963 | 0.7223133247224964 | 1 | 502 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 2444210 | 217166 | NR1D1_MOUSE | Q3UV55 | |
| NR1H2_MOUSE.H10MO.D |  |
Nr1h2 | 19 | D | TMRAGGTCAAAGGTCAASK | HOCOMOCO v9 | 24 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 1352463 | 22260 | NR1H2_MOUSE | Q60644 | ||||
| NR1H4_MOUSE.H10MO.C |  |
Nr1h4 | 14 | C | RRGGTCAnKRWCCY | HOCOMOCO v9 | 45 | Thyroid hormone receptor-related factors (NR1){2.1.2} | LXR (NR1H){2.1.2.7} | 1352464 | 20186 | NR1H4_MOUSE | Q60641 | ||||
| NR1I2_MOUSE.H10MO.C |  |
Nr1i2 | 19 | C | ddRRSTYARnnRAGKTCAb | HOCOMOCO v9 | 29 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 1337040 | 18171 | NR1I2_MOUSE | O54915 | ||||
| NR1I2_MOUSE.H10MO.S |  |
Nr1i2 | 8 | S | dAGKTCAb | HOCOMOCO v9 | 45 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 1337040 | 18171 | NR1I2_MOUSE | O54915 | Secondary motif. | |||
| NR1I3_MOUSE.H10MO.C |  |
Nr1i3 | 18 | C | RRGSTCAvvvRAGKWYRd | HOCOMOCO v9 | 28 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 1346307 | 12355 | NR1I3_MOUSE | O35627 | ||||
| NR1I3_MOUSE.H10MO.S |  |
Nr1i3 | 8 | S | RAGTTCAb | HOCOMOCO v9 | 33 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 1346307 | 12355 | NR1I3_MOUSE | O35627 | Secondary motif. | |||
| NR2C1_MOUSE.H10MO.C |  |
Nr2c1 | 13 | C | RbYvARAGGTCAn | HOCOMOCO v9 | 19 | RXR-related receptors (NR2){2.1.3} | Testicular receptors (NR2C){2.1.3.4} | 1352465 | 22025 | NR2C1_MOUSE | Q505F1 | ||||
| NR2C2_MOUSE.H10MO.A |  |
Nr2c2 | 13 | A | RRSbSARAGGKMR | HOCOMOCO v9 | 2062 | RXR-related receptors (NR2){2.1.3} | Testicular receptors (NR2C){2.1.3.4} | 1352466 | 22026 | NR2C2_MOUSE | P49117 | ||||
| NR2E3_MOUSE.H10MO.C |  |
Nr2e3 | 14 | C | nAAKTCAAAARKMM | HOCOMOCO v9 | 23 | RXR-related receptors (NR2){2.1.3} | Tailless-like receptors (NR2E){2.1.3.3} | 1346317 | 23958 | NR2E3_MOUSE | Q9QXZ7 | ||||
| NR2F6_MOUSE.H10MO.D |  |
Nr2f6 | 18 | D | RGSWMAAAGdnSAnnKdR | HOCOMOCO v9 | 11 | RXR-related receptors (NR2){2.1.3} | COUP-like receptors (NR2F){2.1.3.5} | 1352453 | 13864 | NR2F6_MOUSE | P43136 | ||||
| NR4A1_MOUSE.H10MO.C |  |
Nr4a1 | 9 | C | RAAGGTCAv | HOCOMOCO v9 | 22 | NGFI-B-related receptors (NR4){2.1.4} | NGFI-B (Nur77, NR4A1){2.1.4.0.1} | 1352454 | 15370 | NR4A1_MOUSE | P12813 | ||||
| NR4A2_MOUSE.H10MO.C |  |
Nr4a2 | 9 | C | RAAGGTCAv | HOCOMOCO v9 | 33 | NGFI-B-related receptors (NR4){2.1.4} | NURR1 (NR4A2){2.1.4.0.2} | 1352456 | 18227 | NR4A2_MOUSE | Q06219 | ||||
| NR4A3_MOUSE.H10MO.D |  |
Nr4a3 | 10 | D | CAAAGGTCAS | HOCOMOCO v9 | 5 | NGFI-B-related receptors (NR4){2.1.4} | NOR1 (NR4A3, TEC){2.1.4.0.3} | 1352457 | 18124 | NR4A3_MOUSE | Q9QZB6 | ||||
| NR5A2_MOUSE.H10MO.A |  |
Nr5a2 | 11 | A | bYCAAGGYCRn | ChIP-Seq | 0.9059177435267767 | 0.9111730299697037 | 2 | 500 | FTZ-F1-related receptors (NR5){2.1.5} | LRH-1 (NR5A2){2.1.5.0.2} | 1346834 | 26424 | NR5A2_MOUSE | P45448 | |
| NR6A1_MOUSE.H10MO.B |  |
Nr6a1 | 13 | B | RAGKTCAAGKTCA | HOCOMOCO v9 | 51 | GCNF-related receptors (NR6){2.1.6} | GCNF (NR6A1){2.1.6.0.1} | 1352459 | 14536 | NR6A1_MOUSE | Q64249 | ||||
| NRF1_MOUSE.H10MO.A |  |
Nrf1 | 12 | A | bYGCGCvTGCKY | HOCOMOCO v9 | 1853 | NRF{0.0.6} | NRF-1 (alpha-pal){0.0.6.0.1} | 1332235 | 18181 | NRF1_MOUSE | Q9WU00 | ||||
| OLIG2_MOUSE.H10MO.B |  |
Olig2 | 20 | B | vMCAKCTGbbnnShdbvhdb | ChIP-Seq | 0.7955124233144746 | 0.8161175869990306 | 2 | 500 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1355331 | 50913 | OLIG2_MOUSE | Q9EQW6 | |
| ONEC2_MOUSE.H10MO.D |  |
Onecut2 | 21 | D | RKvhYdTbATKGATTWTWTTW | HOCOMOCO v9 | 7 | HD-CUT factors{3.1.9} | ONECUT{3.1.9.1} | 1891408 | 225631 | ONEC2_MOUSE | Q6XBJ3 | ||||
| OTX1_MOUSE.H10MO.D |  |
Otx1 | 8 | D | nGGATTAn | HOCOMOCO v9 | 8 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 97450 | 18423 | OTX1_MOUSE | P80205 | ||||
| OTX2_MOUSE.H10MO.C |  |
Otx2 | 15 | C | dhdMhvSGATTARvd | HOCOMOCO v9 | 25 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 97451 | OTX2_MOUSE | P80206 | |||||
| OVOL1_MOUSE.H10MO.C |  |
Ovol1 | 9 | C | KGTAACKGT | HOCOMOCO v9 | 7 | More than 3 adjacent zinc finger factors{2.3.3} | OVOL-factors{2.3.3.17} | 1330290 | 18426 | OVOL1_MOUSE | Q9WTJ2 | ||||
| P53_MOUSE.H10MO.C |  |
Tp53 | 20 | C | RRRCAWGYCYvRRCWWGYhb | HOCOMOCO v9 | 1754 | p53-related factors{6.3.1} | p53{6.3.1.0.1} | 98834 | 22059 | P53_MOUSE | P02340 | ||||
| P63_MOUSE.H10MO.D |  |
Tp63 | 11 | D | RRRCWTGTCYn | HOCOMOCO v9 | 22 | p53-related factors{6.3.1} | p63{6.3.1.0.2} | 1330810 | 22061 | P63_MOUSE | O88898 | ||||
| P73_MOUSE.H10MO.D |  |
Tp73 | 12 | D | nRKGCAdGTYbb | HOCOMOCO v9 | 15 | p53-related factors{6.3.1} | p73{6.3.1.0.3} | 1336991 | 22062 | P73_MOUSE | Q9JJP2 | ||||
| PAX2_MOUSE.H10MO.D |  |
Pax2 | 18 | D | vhbnvdTSAhRvdYGAYW | HOCOMOCO v9 | 50 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97486 | 18504 | PAX2_MOUSE | P32114 | ||||
| PAX2_MOUSE.H10MO.S |  |
Pax2 | 7 | S | SCRTGAM | HOCOMOCO v9 | 58 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97486 | 18504 | PAX2_MOUSE | P32114 | Secondary motif. | |||
| PAX5_MOUSE.H10MO.D |  |
Pax5 | 19 | D | vdRnbYMvYvRRGYRKRRM | HOCOMOCO v9 | 4053 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97489 | 18507 | PAX5_MOUSE | Q02650 | ||||
| PAX5_MOUSE.H10MO.S |  |
Pax5 | 8 | S | RSCTGAGb | HOCOMOCO v9 | 3924 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97489 | 18507 | PAX5_MOUSE | Q02650 | Secondary motif. | |||
| PAX6_MOUSE.H10MO.D |  |
Pax6 | 11 | D | TSAdGYGTRAM | HOCOMOCO v9 | 165 | Paired plus homeo domain{3.2.1} | PAX-4/6{3.2.1.2} | 97490 | 18508 | PAX6_MOUSE | P63015 | ||||
| PAX8_MOUSE.H10MO.D |  |
Pax8 | 14 | D | bYvAYYSRvSYRKd | HOCOMOCO v9 | 37 | Paired domain only{3.2.2} | PAX-2-like factors (partial homeobox){3.2.2.2} | 97492 | 18510 | PAX8_MOUSE | Q00288 | ||||
| PBX1_MOUSE.H10MO.B |  |
Pbx1 | 15 | B | ddhWWGATKGATbdb | HOCOMOCO v9 | 145 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 97495 | 18514 | PBX1_MOUSE | P41778 | ||||
| PBX2_MOUSE.H10MO.C |  |
Pbx2 | 15 | C | KvMhKTGATTGWWKb | HOCOMOCO v9 | 19 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 1341793 | 18515 | PBX2_MOUSE | O35984 | ||||
| PBX3_MOUSE.H10MO.A |  |
Pbx3 | 14 | A | bbbYSATTGGYddv | HOCOMOCO v9 | 1885 | TALE-type homeo domain factors{3.1.4} | PBX{3.1.4.4} | 97496 | 18516 | PBX3_MOUSE | O35317 | ||||
| PDX1_MOUSE.H10MO.C |  |
Pdx1 | 9 | C | nSTMATTAR | HOCOMOCO v9 | 74 | HOX-related factors{3.1.1} | PDX{3.1.1.15} | 102851 | 18609 | PDX1_MOUSE | P52946 | ||||
| PEBB_MOUSE.H10MO.C |  |
Cbfb | 11 | C | YbTGTGGTYdb | HOCOMOCO v9 | 23 | 99851 | 12400 | PEBB_MOUSE | Q08024 | ||||||
| PIT1_MOUSE.H10MO.C |  |
Pou1f1 | 13 | C | bWMWTGvAWATWh | HOCOMOCO v9 | 39 | POU domain factors{3.1.10} | POU1 (Pit-1-like factors){3.1.10.1} | 97588 | 18736 | PIT1_MOUSE | Q00286 | ||||
| PITX2_MOUSE.H10MO.D |  |
Pitx2 | 10 | D | YKGGATTWvh | HOCOMOCO v9 | 17 | Paired-related HD factors{3.1.3} | PITX{3.1.3.19} | 109340 | 18741 | PITX2_MOUSE | P97474 | ||||
| PKNX1_MOUSE.H10MO.D |  |
Pknox1 | 13 | D | RdvKTGATWGAYK | HOCOMOCO v9 | 10 | TALE-type homeo domain factors{3.1.4} | PKNOX{3.1.4.5} | 1201409 | 18771 | PKNX1_MOUSE | O70477 | ||||
| PLAG1_MOUSE.H10MO.D |  |
Plag1 | 17 | D | RGGGGSMhndRdMGGGK | HOCOMOCO v9 | 25 | More than 3 adjacent zinc finger factors{2.3.3} | PLAG factors{2.3.3.25} | 1891916 | 56711 | PLAG1_MOUSE | Q9QYE0 | ||||
| PLAG1_MOUSE.H10MO.S |  |
Plag1 | 13 | S | RvdRGGGGSnnbv | HOCOMOCO v9 | 25 | More than 3 adjacent zinc finger factors{2.3.3} | PLAG factors{2.3.3.25} | 1891916 | 56711 | PLAG1_MOUSE | Q9QYE0 | Secondary motif. | |||
| PO2F1_MOUSE.H10MO.B |  |
Pou2f1 | 9 | B | hATGYAAAW | HOCOMOCO v9 | 214 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 101898 | 18986 | PO2F1_MOUSE | P25425 | ||||
| PO2F2_MOUSE.H10MO.B |  |
Pou2f2 | 9 | B | YATGCAAAT | HOCOMOCO v9 | 1296 | POU domain factors{3.1.10} | POU2 (Oct-1/2-like factors){3.1.10.2} | 101897 | 18987 | PO2F2_MOUSE | Q00196 | ||||
| PO3F1_MOUSE.H10MO.C |  |
Pou3f1 | 14 | C | vRTKSTWATGCWWd | HOCOMOCO v9 | 16 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 101896 | 18991 | PO3F1_MOUSE | P21952 | ||||
| PO3F2_MOUSE.H10MO.D |  |
Pou3f2 | 10 | D | hWWATTYAdd | HOCOMOCO v9 | 66 | POU domain factors{3.1.10} | POU3 (Oct-6-like factors){3.1.10.3} | 101895 | 18992 | PO3F2_MOUSE | P31360 | ||||
| PO4F2_MOUSE.H10MO.D |  |
Pou4f2 | 13 | D | YvTTAATGAKhnn | HOCOMOCO v9 | 31 | POU domain factors{3.1.10} | POU4 (Brn-3-like factors){3.1.10.4} | 102524 | 18997 | PO4F2_MOUSE | Q63934 | ||||
| PO5F1_MOUSE.H10MO.A |  |
Pou5f1 | 16 | A | bYWTTSWhATGYARAW | HOCOMOCO v9 | 0.8948666484183225 | 0.966759112085938 | 6 | 1396 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 101893 | 18999 | PO5F1_MOUSE | P20263 | |
| PO6F1_MOUSE.H10MO.D |  |
Pou6f1 | 13 | D | nATAATTTATGCR | HOCOMOCO v9 | 14 | POU domain factors{3.1.10} | POU6 (Brn-5-like factors){3.1.10.6} | 102935 | 19009 | PO6F1_MOUSE | Q07916 | ||||
| PPARA_MOUSE.H10MO.C |  |
Ppara | 13 | C | RGGbSAvAGGKCR | HOCOMOCO v9 | 111 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 104740 | 19013 | PPARA_MOUSE | P23204 | ||||
| PPARA_MOUSE.H10MO.S |  |
Ppara | 7 | S | dAGGTCA | HOCOMOCO v9 | 119 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 104740 | 19013 | PPARA_MOUSE | P23204 | Secondary motif. | |||
| PPARD_MOUSE.H10MO.D |  |
Ppard | 14 | D | YAGKnnAMAGGWYA | HOCOMOCO v9 | 18 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 101884 | 19015 | PPARD_MOUSE | P35396 | ||||
| PPARG_MOUSE.H10MO.A |  |
Pparg | 17 | A | hWbWRKGdbARAGGKbR | ChIP-Seq | 0.8256461461519156 | 0.8803082910391935 | 6 | 501 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 97747 | 19016 | PPARG_MOUSE | P37238 | |
| PPARG_MOUSE.H10MO.S | 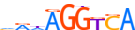 |
Pparg | 9 | S | hRdAGGKCA | HOCOMOCO v9 | 0.6440152618939315 | 0.6655587349852932 | 6 | 1004 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 97747 | 19016 | PPARG_MOUSE | P37238 | Secondary motif. |
| PRD14_MOUSE.H10MO.D |  |
Prdm14 | 12 | D | dSWTAGRGMMYh | ChIP-Seq | 0.777648129450664 | 0.777648129450664 | 1 | 501 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 3588194 | 383491 | PRD14_MOUSE | E9Q3T6 | |
| PRDM1_MOUSE.H10MO.A |  |
Prdm1 | 14 | A | RRAAGKGAAAGKRv | HOCOMOCO v9 | 856 | More than 3 adjacent zinc finger factors{2.3.3} | PRDM1-like factors{2.3.3.12} | 99655 | 12142 | PRDM1_MOUSE | Q60636 | ||||
| PRGR_MOUSE.H10MO.C |  |
Pgr | 13 | C | RGAACAnhbTGYh | HOCOMOCO v9 | 26 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 97567 | PRGR_MOUSE | Q00175 | |||||
| PRGR_MOUSE.H10MO.S |  |
Pgr | 7 | S | RGAACAb | HOCOMOCO v9 | 40 | Steroid hormone receptors (NR3){2.1.1} | GR-like receptors (NR3C){2.1.1.1} | 97567 | PRGR_MOUSE | Q00175 | Secondary motif. | ||||
| PROP1_MOUSE.H10MO.D |  |
Prop1 | 15 | D | nAGnAAWTnAnATRd | HOCOMOCO v9 | 9 | Paired-related HD factors{3.1.3} | PROP{3.1.3.20} | 109330 | 19127 | PROP1_MOUSE | P97458 | ||||
| PRRX1_MOUSE.H10MO.D | 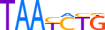 |
Prrx1 | 7 | D | TAATCTG | HOCOMOCO v9 | 4 | Paired-related HD factors{3.1.3} | PRRX{3.1.3.21} | 97712 | 18933 | PRRX1_MOUSE | P63013 | ||||
| PRRX2_MOUSE.H10MO.C |  |
Prrx2 | 7 | C | hTAATTR | HOCOMOCO v9 | 65 | Paired-related HD factors{3.1.3} | PRRX{3.1.3.21} | 98218 | 20204 | PRRX2_MOUSE | Q06348 | ||||
| PTF1A_MOUSE.H10MO.C |  |
Ptf1a | 21 | C | nRRGARAnnbhMCASSTGYCM | HOCOMOCO v9 | 10 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 1328312 | 19213 | PTF1A_MOUSE | Q9QX98 | ||||
| PURA_MOUSE.H10MO.D |  |
Pura | 17 | D | RGWvKbKGnKGSMRKGK | HOCOMOCO v9 | 13 | PUR{0.0.5} | Pur-alpha (PURA, PUR1){0.0.5.0.1} | 103079 | 19290 | PURA_MOUSE | P42669 | ||||
| RARA_MOUSE.H10MO.C |  |
Rara | 17 | C | RGKTSAnvvdvRSKKSR | HOCOMOCO v9 | 122 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97856 | 19401 | RARA_MOUSE | P11416 | ||||
| RARA_MOUSE.H10MO.S |  |
Rara | 7 | S | vAGGTCA | HOCOMOCO v9 | 77 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97856 | 19401 | RARA_MOUSE | P11416 | Secondary motif. | |||
| RARB_MOUSE.H10MO.D |  |
Rarb | 11 | D | vAGGTCAnvRv | HOCOMOCO v9 | 25 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97857 | 218772 | RARB_MOUSE | P22605 | ||||
| RARG_MOUSE.H10MO.C |  |
Rarg | 20 | C | dddGbdbRvMnvRAGGTCAv | HOCOMOCO v9 | 24 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97858 | 19411 | RARG_MOUSE | P18911 | ||||
| RARG_MOUSE.H10MO.S |  |
Rarg | 7 | S | RAGGTCA | HOCOMOCO v9 | 32 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Retinoic acid receptors (NR1B){2.1.2.1} | 97858 | 19411 | RARG_MOUSE | P18911 | Secondary motif. | |||
| RELB_MOUSE.H10MO.C |  |
Relb | 12 | C | vRGGGMTTTSMh | HOCOMOCO v9 | 28 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 103289 | 19698 | RELB_MOUSE | Q04863 | ||||
| REL_MOUSE.H10MO.C |  |
Rel | 12 | C | dKGGRnWTTCCv | HOCOMOCO v9 | 75 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 97897 | REL_MOUSE | P15307 | |||||
| REST_MOUSE.H10MO.A |  |
Rest | 21 | A | bMRSSWMYdhGGMCAGMdMMh | ChIP-Seq | 0.8789557760012597 | 0.9118631451175175 | 2 | 431 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 104897 | 19712 | REST_MOUSE | Q8VIG1 | |
| RFX1_MOUSE.H10MO.C |  |
Rfx1 | 22 | C | bbbbSbGTTGCCWdGGvvMSvv | ChIP-Seq | 0.8886301038522996 | 0.8886301038522996 | 1 | 457 | RFX-related factors{3.3.3} | RFX1 (EF-C){3.3.3.0.1} | 105982 | 19724 | RFX1_MOUSE | P48377 | |
| RFX2_MOUSE.H10MO.C |  |
Rfx2 | 13 | C | dGTTGCYMRGSAR | HOCOMOCO v9 | 13 | RFX-related factors{3.3.3} | RFX2{3.3.3.0.2} | 106583 | 19725 | RFX2_MOUSE | P48379 | ||||
| RFX3_MOUSE.H10MO.B |  |
Rfx3 | 14 | B | bGTTRCCATGGhRn | HOCOMOCO v9 | 772 | RFX-related factors{3.3.3} | RFX3{3.3.3.0.3} | 106582 | 19726 | RFX3_MOUSE | P48381 | ||||
| RORA_MOUSE.H10MO.B |  |
Rora | 12 | B | dWAAbTAGGTCA | HOCOMOCO v9 | 115 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 104661 | 19883 | RORA_MOUSE | P51448 | ||||
| RORG_MOUSE.H10MO.C |  |
Rorc | 13 | C | nAAWvhAGGTCAG | HOCOMOCO v9 | 7 | Thyroid hormone receptor-related factors (NR1){2.1.2} | ROR (NR1F){2.1.2.6} | 104856 | 19885 | RORG_MOUSE | P51450 | ||||
| RREB1_MOUSE.H10MO.D |  |
Rreb1 | 22 | D | dbSKbdGKKGKTGbTKKGKGGd | HOCOMOCO v9 | 26 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 2443664 | 68750 | RREB1_MOUSE | Q3UH06 | ||||
| RUNX1_MOUSE.H10MO.B |  |
Runx1 | 10 | B | dbTGYGGThd | HOCOMOCO v9 | 0.7789709633569243 | 0.7789709633569243 | 1 | 86 | Runt-related factors{6.4.1} | Runx1 (PEBP2alphaB, CBF-alpha2, AML-1){6.4.1.0.2} | 99852 | RUNX1_MOUSE | Q03347 | ||
| RUNX2_MOUSE.H10MO.B |  |
Runx2 | 11 | B | bTGTGGTKdbb | HOCOMOCO v9 | 73 | Runt-related factors{6.4.1} | Runx2 (PEBP2alphaA, CBF-alpha1){6.4.1.0.1} | 99829 | 12393 | RUNX2_MOUSE | Q08775 | ||||
| RUNX3_MOUSE.H10MO.B |  |
Runx3 | 14 | B | bKKKKTTTGYGGTb | HOCOMOCO v9 | 284 | Runt-related factors{6.4.1} | Runx3 (PEBP2alphaC, CBF-alpha3, AML-2){6.4.1.0.3} | 102672 | RUNX3_MOUSE | Q64131 | |||||
| RXRA_MOUSE.H10MO.C |  |
Rxra | 22 | C | dRddbhvdRAGKTCAAGGbSRb | ChIP-Seq | 0.7491064014711164 | 0.7811442288632731 | 3 | 500 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98214 | 20181 | RXRA_MOUSE | P28700 | |
| RXRB_MOUSE.H10MO.C |  |
Rxrb | 10 | C | WSAGGTCAbd | HOCOMOCO v9 | 30 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98215 | 20182 | RXRB_MOUSE | P28704 | ||||
| RXRG_MOUSE.H10MO.C |  |
Rxrg | 17 | C | RAGKTCAAGGhYRbbbY | ChIP-Seq | 0.848851531419049 | 0.848851531419049 | 1 | 509 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98216 | 20183 | RXRG_MOUSE | P28705 | |
| SMAD1_MOUSE.H10MO.D | 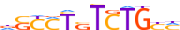 |
Smad1 | 12 | D | dGCCTSTCTGYM | HOCOMOCO v9 | 12 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 109452 | 17125 | SMAD1_MOUSE | P70340 | ||||
| SMAD2_MOUSE.H10MO.D |  |
Smad2 | 11 | D | KKGKCYGKMTv | HOCOMOCO v9 | 18 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 108051 | 17126 | SMAD2_MOUSE | Q62432 | ||||
| SMAD3_MOUSE.H10MO.C |  |
Smad3 | 10 | C | vhGTCTGbvb | HOCOMOCO v9 | 0.6549951843899126 | 0.6549951843899126 | 1 | 74 | SMAD factors{7.1.1} | Regulatory Smads (R-Smad){7.1.1.1} | 1201674 | 17127 | SMAD3_MOUSE | Q8BUN5 | |
| SMAD4_MOUSE.H10MO.C |  |
Smad4 | 9 | C | WGTCTGbMh | HOCOMOCO v9 | 81 | SMAD factors{7.1.1} | Co-activating Smads (Co-Smad){7.1.1.2} | 894293 | 17128 | SMAD4_MOUSE | P97471 | ||||
| SMRC1_MOUSE.H10MO.B |  |
Smarcc1 | 9 | B | vTGAGTCAb | HOCOMOCO v9 | 2119 | Myb/SANT domain factors{3.5.1} | TADA2-like factors{3.5.1.6} | 1203524 | 20588 | SMRC1_MOUSE | P97496 | ||||
| SNAI1_MOUSE.H10MO.C |  |
Snai1 | 8 | C | vCAGGTGb | HOCOMOCO v9 | 5 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 98330 | 20613 | SNAI1_MOUSE | Q02085 | ||||
| SNAI2_MOUSE.H10MO.C |  |
Snai2 | 7 | C | nCAGGTG | HOCOMOCO v9 | 6 | More than 3 adjacent zinc finger factors{2.3.3} | Snail-like factors{2.3.3.2} | 1096393 | 20583 | SNAI2_MOUSE | P97469 | ||||
| SOX10_MOUSE.H10MO.D |  |
Sox10 | 7 | D | bCWTTGT | HOCOMOCO v9 | 32 | 98358 | 20665 | SOX10_MOUSE | Q04888 | ||||||
| SOX13_MOUSE.H10MO.D |  |
Sox13 | 8 | D | nATTGTKn | HOCOMOCO v9 | 5 | SOX-related factors{4.1.1} | Group D{4.1.1.4} | 98361 | 20668 | SOX13_MOUSE | Q04891 | ||||
| SOX15_MOUSE.H10MO.D |  |
Sox15 | 7 | D | CWTTGTT | HOCOMOCO v9 | 8 | SOX-related factors{4.1.1} | Group G{4.1.1.7} | 98363 | 20670 | SOX15_MOUSE | P43267 | ||||
| SOX17_MOUSE.H10MO.D |  |
Sox17 | 18 | D | MAMCAAWnhYYATTGTSv | HOCOMOCO v9 | 57 | SOX-related factors{4.1.1} | Group F{4.1.1.6} | 107543 | 20671 | SOX17_MOUSE | Q61473 | ||||
| SOX18_MOUSE.H10MO.D |  |
Sox18 | 19 | D | SMWhnMATTGTYbWnnYYC | HOCOMOCO v9 | 8 | SOX-related factors{4.1.1} | Group F{4.1.1.6} | 103559 | 20672 | SOX18_MOUSE | P43680 | ||||
| SOX2_MOUSE.H10MO.A | 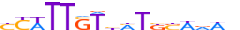 |
Sox2 | 16 | A | YYWTTSTbMTKSWdWh | HOCOMOCO v9 | 0.929980421199365 | 0.9777416312648883 | 5 | 678 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 98364 | 20674 | SOX2_MOUSE | P48432 | |
| SOX3_MOUSE.H10MO.C |  |
Sox3 | 11 | C | bSCTTTGTYYb | ChIP-Seq | 0.8639200352740097 | 0.8639200352740097 | 1 | 500 | SOX-related factors{4.1.1} | Group B{4.1.1.2} | 98365 | SOX3_MOUSE | P53784 | ||
| SOX4_MOUSE.H10MO.C |  |
Sox4 | 12 | C | CSCTTTGTTYTn | HOCOMOCO v9 | 8 | SOX-related factors{4.1.1} | Group C{4.1.1.3} | 98366 | 20677 | SOX4_MOUSE | Q06831 | ||||
| SOX5_MOUSE.H10MO.C |  |
Sox5 | 8 | C | bATTGTTh | HOCOMOCO v9 | 69 | SOX-related factors{4.1.1} | Group D{4.1.1.4} | 98367 | 20678 | SOX5_MOUSE | P35710 | ||||
| SOX9_MOUSE.H10MO.B |  |
Sox9 | 11 | B | bYYATTGTTYh | HOCOMOCO v9 | 158 | SOX-related factors{4.1.1} | Group E{4.1.1.5} | 98371 | 20682 | SOX9_MOUSE | Q04887 | ||||
| SP1_MOUSE.H10MO.A | 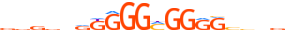 |
Sp1 | 19 | A | vvvvvvKGGGMGGRRvbnv | HOCOMOCO v9 | 3064 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 98372 | 20683 | SP1_MOUSE | O89090 | ||||
| SP1_MOUSE.H10MO.S |  |
Sp1 | 11 | S | RKGGGMGGRRv | HOCOMOCO v9 | 3614 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 98372 | 20683 | SP1_MOUSE | O89090 | Secondary motif. | |||
| SP2_MOUSE.H10MO.C |  |
Sp2 | 17 | C | ddRKRRGGSGKRRbbRv | HOCOMOCO v9 | 31 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 1926162 | 78912 | SP2_MOUSE | Q9D2H6 | ||||
| SP3_MOUSE.H10MO.B |  |
Sp3 | 20 | B | vvvvvKGGGMGGRSvbdvvv | HOCOMOCO v9 | 415 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 1277166 | 20687 | SP3_MOUSE | O70494 | ||||
| SP4_MOUSE.H10MO.D |  |
Sp4 | 24 | D | RSRdSSRKGGSSGGSGSGSvGbSv | HOCOMOCO v9 | 23 | Three-zinc finger Krüppel-related factors{2.3.1} | Sp1-like factors{2.3.1.1} | 107595 | SP4_MOUSE | Q62445 | |||||
| SPI1_MOUSE.H10MO.A |  |
Spi1 | 16 | A | dvWAWKRGGAARTGvv | ChIP-Seq | 0.9546360121380553 | 0.982990458166321 | 7 | 493 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 98282 | 20375 | SPI1_MOUSE | P17433 | |
| SPIB_MOUSE.H10MO.B |  |
Spib | 12 | B | vvAAASMGGAAS | HOCOMOCO v9 | 82 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 892986 | 272382 | SPIB_MOUSE | O35906 | ||||
| SPZ1_MOUSE.H10MO.D |  |
Spz1 | 16 | D | nSRSKKTTMCMbhbGn | HOCOMOCO v9 | 11 | 1930801 | 79401 | SPZ1_MOUSE | Q99MY0 | ||||||
| SRBP1_MOUSE.H10MO.B |  |
Srebf1 | 11 | B | vRTSRSSWGWb | HOCOMOCO v9 | 2191 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 107606 | 20787 | SRBP1_MOUSE | Q9WTN3 | ||||
| SRBP2_MOUSE.H10MO.B |  |
Srebf2 | 13 | B | vvvWGGvSWGRnb | HOCOMOCO v9 | 2363 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 107585 | 20788 | SRBP2_MOUSE | Q3U1N2 | ||||
| SRF_MOUSE.H10MO.A |  |
Srf | 15 | A | hWKbCCWWATAWGGh | HOCOMOCO v9 | 2187 | Responders to external signals (SRF/RLM1){5.1.2} | SRF{5.1.2.0.1} | 106658 | 20807 | SRF_MOUSE | Q9JM73 | ||||
| SRY_MOUSE.H10MO.B |  |
Sry | 9 | B | bWTTGTWWh | HOCOMOCO v9 | 108 | SOX-related factors{4.1.1} | Group A{4.1.1.1} | 98660 | 21674 | SRY_MOUSE | Q05738 | ||||
| STA5A_MOUSE.H10MO.A |  |
Stat5a | 13 | A | ddTTCYTRGAAdh | ChIP-Seq | 0.9165148421625577 | 0.9596678321611117 | 6 | 500 | STAT factors{6.2.1} | STAT5A{6.2.1.0.5} | 103036 | 20850 | STA5A_MOUSE | P42230 | |
| STA5B_MOUSE.H10MO.C |  |
Stat5b | 11 | C | ndTTCYTRGAA | ChIP-Seq | 0.94228813905837 | 0.94228813905837 | 1 | 500 | STAT factors{6.2.1} | STAT5B{6.2.1.0.6} | 103035 | 20851 | STA5B_MOUSE | P42232 | |
| STAT1_MOUSE.H10MO.A |  |
Stat1 | 19 | A | bhYYYdSTTYChnWTTYhb | ChIP-Seq | 0.790531917702979 | 0.8509351045395153 | 6 | 516 | STAT factors{6.2.1} | STAT1{6.2.1.0.1} | 103063 | STAT1_MOUSE | P42225 | ||
| STAT2_MOUSE.H10MO.B |  |
Stat2 | 15 | B | dhRSTTTCnbTTYYh | HOCOMOCO v9 | 2025 | STAT factors{6.2.1} | STAT2{6.2.1.0.2} | 103039 | STAT2_MOUSE | Q9WVL2 | |||||
| STAT3_MOUSE.H10MO.B |  |
Stat3 | 15 | B | bbbYvYTTYCYRKvA | ChIP-Seq | 0.7752409526996936 | 0.8457593740459493 | 4 | 499 | STAT factors{6.2.1} | STAT3{6.2.1.0.3} | 103038 | 20848 | STAT3_MOUSE | P42227 | |
| STAT4_MOUSE.H10MO.A |  |
Stat4 | 11 | A | hTTCYTdKAAv | ChIP-Seq | 0.8494972594919306 | 0.8735780469016585 | 2 | 500 | STAT factors{6.2.1} | STAT4{6.2.1.0.4} | 103062 | 20849 | STAT4_MOUSE | P42228 | |
| STAT6_MOUSE.H10MO.C |  |
Stat6 | 12 | C | dbTTCYWvRGAA | ChIP-Seq | 0.8131608397366931 | 0.8131608397366931 | 1 | 501 | STAT factors{6.2.1} | STAT6{6.2.1.0.7} | 103034 | 20852 | STAT6_MOUSE | P52633 | |
| STF1_MOUSE.H10MO.B |  |
Nr5a1 | 9 | B | hSAAGGhYR | HOCOMOCO v9 | 120 | FTZ-F1-related receptors (NR5){2.1.5} | FTZ-F1 (SF-1) (NR5A1){2.1.5.0.1} | 1346833 | 26423 | STF1_MOUSE | P33242 | ||||
| SUH_MOUSE.H10MO.C |  |
Rbpj | 9 | C | hRTGGGAAM | HOCOMOCO v9 | 65 | CSL-related factors{6.1.4} | M{6.1.4.1} | 96522 | 19664 | SUH_MOUSE | P31266 | ||||
| TAL1_MOUSE.H10MO.A | 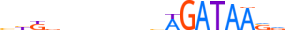 |
Tal1 | 20 | A | bbKbnnnnndvAGATAASvv | ChIP-Seq | 0.9065256501998835 | 0.9600775252896189 | 11 | 500 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 98480 | 21349 | TAL1_MOUSE | P22091 | |
| TAL1_MOUSE.H10MO.S |  |
Tal1 | 12 | S | vMMCAGATGGTn | HOCOMOCO v9 | 0.5368454580087119 | 0.5749838743437456 | 11 | 47 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 98480 | 21349 | TAL1_MOUSE | P22091 | Secondary motif. |
| TBP_MOUSE.H10MO.C |  |
Tbp | 11 | C | vvKATAWAWdv | HOCOMOCO v9 | 102 | TBP-related factors{8.1.1} | TBP{8.1.1.0.1} | 101838 | 21374 | TBP_MOUSE | P29037 | ||||
| TBX20_MOUSE.H10MO.C |  |
Tbx20 | 13 | C | vdSTGnTGACARv | ChIP-Seq | 0.8085061772309449 | 0.808506177230945 | 1 | 500 | TBX1-related factors{6.5.3} | TBX20{6.5.3.0.5} | 1888496 | 57246 | TBX20_MOUSE | Q9ES03 | |
| TBX2_MOUSE.H10MO.D |  |
Tbx2 | 25 | D | hbMhWhChbAGGTGTGAnRWKnSMn | HOCOMOCO v9 | 9 | TBX2-related factors{6.5.4} | TBX2{6.5.4.0.1} | 98494 | 21385 | TBX2_MOUSE | Q60707 | ||||
| TBX3_MOUSE.H10MO.D |  |
Tbx3 | 24 | D | MRKWWGRSdnYYAGGTGATAARWM | HOCOMOCO v9 | 10 | TBX2-related factors{6.5.4} | TBX3{6.5.4.0.2} | 98495 | 21386 | TBX3_MOUSE | P70324 | ||||
| TBX5_MOUSE.H10MO.D |  |
Tbx5 | 8 | D | ARGTGKSW | HOCOMOCO v9 | 38 | TBX2-related factors{6.5.4} | TBX5{6.5.4.0.4} | 102541 | 21388 | TBX5_MOUSE | P70326 | ||||
| TCF7_MOUSE.H10MO.C |  |
Tcf7 | 12 | C | RSWWCAAAGSdb | HOCOMOCO v9 | 16 | TCF-7-related factors{4.1.3} | TCF-7 (TCF-1) [1]{4.1.3.0.1} | 98507 | 21414 | TCF7_MOUSE | Q00417 | ||||
| TEAD1_MOUSE.H10MO.D |  |
Tead1 | 14 | D | bRMATTYYWShdbh | HOCOMOCO v9 | 30 | TEF-1-related factors{3.6.1} | TEF-1 (TEAD-1, TCF-13){3.6.1.0.1} | 101876 | 21676 | TEAD1_MOUSE | P30051 | ||||
| TEAD3_MOUSE.H10MO.D |  |
Tead3 | 15 | D | nAWATTYCTGhhbYh | HOCOMOCO v9 | 12 | TEF-1-related factors{3.6.1} | TEF-5 (TEAD-3, TEAD5){3.6.1.0.4} | 109241 | 21678 | TEAD3_MOUSE | P70210 | ||||
| TEAD4_MOUSE.H10MO.D |  |
Tead4 | 12 | D | AAWAATAKCYCW | HOCOMOCO v9 | 11 | TEF-1-related factors{3.6.1} | TEF-3 (TEAD-4, TCF-13L1, Tefr1){3.6.1.0.2} | 106907 | 21679 | TEAD4_MOUSE | Q62296 | ||||
| TEF_MOUSE.H10MO.D |  |
Tef | 14 | D | TGdTKAYRTMdvYd | HOCOMOCO v9 | 14 | C/EBP-related{1.1.8} | PAR factors{1.1.8.2} | 98663 | 21685 | TEF_MOUSE | Q9JLC6 | ||||
| TF2L1_MOUSE.H10MO.C |  |
Tfcp2l1 | 20 | C | dRdCYRGhhdRARCYRGhhb | ChIP-Seq | 0.9527412135801124 | 0.9527412135801123 | 1 | 467 | CP2-related factors{6.7.2} | CP2-L1 (LBP-9, CRTR-1){6.7.2.0.2} | 2444691 | 81879 | TF2L1_MOUSE | Q3UNW5 | |
| TF65_MOUSE.H10MO.C | 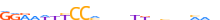 |
Rela | 22 | C | RRRRWKYCCMnnvYKdbnhMWb | ChIP-Seq | 0.8139897406768539 | 0.8139897406768538 | 1 | 500 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 103290 | 19697 | TF65_MOUSE | Q04207 | |
| TF65_MOUSE.H10MO.S |  |
Rela | 11 | S | KGGRvTTTCCv | HOCOMOCO v9 | 0.7793487326912545 | 0.7793487326912545 | 1 | 2165 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 103290 | 19697 | TF65_MOUSE | Q04207 | Secondary motif. |
| TF7L2_MOUSE.H10MO.A |  |
Tcf7l2 | 12 | A | bbCTTTGWWbbb | HOCOMOCO v9 | 1316 | TCF-7-related factors{4.1.3} | TCF-7L2 (TCF-4) [1]{4.1.3.0.3} | 1202879 | 21416 | TF7L2_MOUSE | Q924A0 | ||||
| TFCP2_MOUSE.H10MO.D |  |
Tfcp2 | 16 | D | SSCWGvnMdvRCMRGM | HOCOMOCO v9 | 13 | CP2-related factors{6.7.2} | CP2 (LSF, SEF){6.7.2.0.1} | 98509 | 21422 | TFCP2_MOUSE | Q9ERA0 | ||||
| TFDP1_MOUSE.H10MO.D |  |
Tfdp1 | 14 | D | RWWKRGCGGGAAAY | HOCOMOCO v9 | 7 | E2F-related factors{3.3.2} | Dp-1{3.3.2.2} | 101934 | 21781 | TFDP1_MOUSE | Q08639 | ||||
| TFE2_MOUSE.H10MO.A | 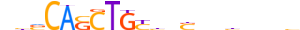 |
Tcf3 | 20 | A | bbvCASCTGYbbShbbbbbb | ChIP-Seq | 0.8526179570043664 | 0.8616653706548376 | 3 | 501 | E2A-related factors{1.2.1} | E2A (TCF-3, ITF-1){1.2.1.0.1} | 98510 | 21423 | TFE2_MOUSE | P15806 | |
| TFE2_MOUSE.H10MO.S |  |
Tcf3 | 11 | S | dMCAGRTGbYb | HOCOMOCO v9 | 0.6847457954180287 | 0.7156127759186904 | 3 | 142 | E2A-related factors{1.2.1} | E2A (TCF-3, ITF-1){1.2.1.0.1} | 98510 | 21423 | TFE2_MOUSE | P15806 | Secondary motif. |
| TFE3_MOUSE.H10MO.C | 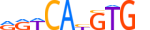 |
Tfe3 | 10 | C | RRWCAYGTGv | HOCOMOCO v9 | 17 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 98511 | 209446 | TFE3_MOUSE | Q64092 | ||||
| TFEB_MOUSE.H10MO.C | 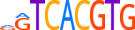 |
Tfeb | 9 | C | dGTCACGTG | HOCOMOCO v9 | 301 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 103270 | 21425 | TFEB_MOUSE | Q9R210 | ||||
| TGIF1_MOUSE.H10MO.D |  |
Tgif1 | 16 | D | vnvnTSWCAnnTSMCn | HOCOMOCO v9 | 14 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 1194497 | 21815 | TGIF1_MOUSE | P70284 | ||||
| TGIF1_MOUSE.H10MO.S |  |
Tgif1 | 7 | S | nTGACAR | HOCOMOCO v9 | 18 | TALE-type homeo domain factors{3.1.4} | TGIF{3.1.4.6} | 1194497 | 21815 | TGIF1_MOUSE | P70284 | Secondary motif. | |||
| THA_MOUSE.H10MO.C | 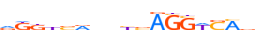 |
Thra | 17 | C | dKKhvRnhbvAGGWSMb | HOCOMOCO v9 | 75 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 98742 | 21833 | THA_MOUSE | P63058 | ||||
| THA_MOUSE.H10MO.S |  |
Thra | 10 | S | nYvAGGTCAv | HOCOMOCO v9 | 76 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 98742 | 21833 | THA_MOUSE | P63058 | Secondary motif. | |||
| THB_MOUSE.H10MO.C | 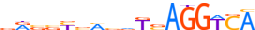 |
Thrb | 17 | C | vRvSYvMbvKSAGGTCA | HOCOMOCO v9 | 38 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 98743 | 21834 | THB_MOUSE | P37242 | ||||
| THB_MOUSE.H10MO.S |  |
Thrb | 10 | S | bvTSAGGTCA | HOCOMOCO v9 | 58 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Thyroid hormone receptors (NR1A){2.1.2.2} | 98743 | 21834 | THB_MOUSE | P37242 | Secondary motif. | |||
| TLX1_MOUSE.H10MO.D |  |
Tlx1 | 17 | D | hYKKYAvbndvMYnRKd | HOCOMOCO v9 | 50 | NK-related factors{3.1.2} | TLX{3.1.2.21} | 98769 | TLX1_MOUSE | P43345 | |||||
| TLX1_MOUSE.H10MO.S |  |
Tlx1 | 11 | S | bGSYAAKddGb | HOCOMOCO v9 | 51 | NK-related factors{3.1.2} | TLX{3.1.2.21} | 98769 | TLX1_MOUSE | P43345 | Secondary motif. | ||||
| TWST1_MOUSE.H10MO.D | 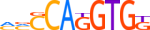 |
Twist1 | 10 | D | hMCCAGGTGG | HOCOMOCO v9 | 5 | Tal-related factors{1.2.3} | Twist-like factors{1.2.3.2} | 98872 | 22160 | TWST1_MOUSE | P26687 | ||||
| TYY1_MOUSE.H10MO.A |  |
Yy1 | 12 | A | vAAvATGGMbRM | HOCOMOCO v9 | 0.8520452390280783 | 0.9271220640513054 | 3 | 1852 | More than 3 adjacent zinc finger factors{2.3.3} | YY1-like factors{2.3.3.9} | 99150 | 22632 | TYY1_MOUSE | Q00899 | |
| UBIP1_MOUSE.H10MO.D |  |
Ubp1 | 7 | D | nCAGAdA | HOCOMOCO v9 | 5 | CP2-related factors{6.7.2} | UBP-1 (LBP-1, SEF, CP2b){6.7.2.0.3} | 104889 | 22221 | UBIP1_MOUSE | Q811S7 | ||||
| USF1_MOUSE.H10MO.A |  |
Usf1 | 10 | A | RGTCACGTGv | HOCOMOCO v9 | 0.9807281325118212 | 0.9853376601206956 | 2 | 2294 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 99542 | 22278 | USF1_MOUSE | Q61069 | |
| USF2_MOUSE.H10MO.A |  |
Usf2 | 13 | A | dGTCACGTGvbbv | ChIP-Seq | 0.9798761500386943 | 0.9890098252519984 | 2 | 324 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 99961 | 22282 | USF2_MOUSE | Q64705 | |
| VDR_MOUSE.H10MO.C |  |
Vdr | 16 | C | RGGTYAhnRRGKKSRb | HOCOMOCO v9 | 115 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 103076 | 22337 | VDR_MOUSE | P48281 | ||||
| VDR_MOUSE.H10MO.S |  |
Vdr | 7 | S | RRGKTCA | HOCOMOCO v9 | 126 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Vitamin D receptor (NR1I){2.1.2.4} | 103076 | 22337 | VDR_MOUSE | P48281 | Secondary motif. | |||
| VSX2_MOUSE.H10MO.D |  |
Vsx2 | 11 | D | TAATTAGCnhd | HOCOMOCO v9 | 23 | Paired-related HD factors{3.1.3} | VSX{3.1.3.28} | 88401 | 12677 | VSX2_MOUSE | Q61412 | ||||
| WT1_MOUSE.H10MO.D |  |
Wt1 | 13 | D | ShGbGGGYGKRKS | HOCOMOCO v9 | 68 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 98968 | WT1_MOUSE | P22561 | |||||
| XBP1_MOUSE.H10MO.C |  |
Xbp1 | 12 | C | GACGTGTMhhWd | HOCOMOCO v9 | 1910 | XBP-1-related factors{1.1.5} | XBP-1{1.1.5.0.1} | 98970 | 22433 | XBP1_MOUSE | O35426 | ||||
| YBOX1_MOUSE.H10MO.D |  |
Ybx1 | 11 | D | bSbSMTTSSYY | HOCOMOCO v9 | 63 | Dbp factors{9.1.1} | B (DbpB/YB-1-like){9.1.1.2} | 99146 | 22608 | YBOX1_MOUSE | P62960 | ||||
| ZBT18_MOUSE.H10MO.D |  |
Zbtb18 | 14 | D | KCCAGATGTTbvSn | HOCOMOCO v9 | 18 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF238-like factors{2.3.3.16} | 1353609 | 30928 | ZBT18_MOUSE | Q9WUK6 | ||||
| ZBT7A_MOUSE.H10MO.D |  |
Zbtb7a | 17 | D | vKShSKGGKYnbvSbdv | HOCOMOCO v9 | 38 | More than 3 adjacent zinc finger factors{2.3.3} | ZBTB7 factors{2.3.3.8} | 1335091 | 16969 | ZBT7A_MOUSE | O88939 | ||||
| ZBTB6_MOUSE.H10MO.D |  |
Zbtb6 | 13 | D | vGAYSMTRGAGCS | HOCOMOCO v9 | 33 | More than 3 adjacent zinc finger factors{2.3.3} | ZBTB6-like factors{2.3.3.11} | 2442998 | 241322 | ZBTB6_MOUSE | Q8K088 | ||||
| ZEB1_MOUSE.H10MO.D |  |
Zeb1 | 7 | D | nMAGGTG | HOCOMOCO v9 | 126 | HD-ZF factors{3.1.8} | ZEB{3.1.8.3} | 1344313 | 21417 | ZEB1_MOUSE | Q64318 | ||||
| ZEP1_MOUSE.H10MO.D |  |
Hivep1 | 12 | D | KGGGAAATCCCn | HOCOMOCO v9 | 10 | Factors with multiple dispersed zinc fingers{2.3.4} | HIV EP-factors{2.3.4.5} | 96100 | ZEP1_MOUSE | Q03172 | |||||
| ZEP2_MOUSE.H10MO.D |  |
Hivep2 | 14 | D | KSYdbGGWAASnSS | HOCOMOCO v9 | 16 | Factors with multiple dispersed zinc fingers{2.3.4} | HIV EP-factors{2.3.4.5} | 1338076 | 15273 | ZEP2_MOUSE | Q3UHF7 | ||||
| ZFHX3_MOUSE.H10MO.D |  |
Zfhx3 | 11 | D | RTTAATWATTW | HOCOMOCO v9 | 7 | HD-ZF factors{3.1.8} | ZFHX{3.1.8.4} | 99948 | ZFHX3_MOUSE | Q61329 | |||||
| ZFX_MOUSE.H10MO.C |  |
Zfx | 19 | C | vvvvvbnbbAGGCCbvGSh | HOCOMOCO v9 | 0.7425756334923719 | 0.7425756334923719 | 1 | 480 | More than 3 adjacent zinc finger factors{2.3.3} | ZFX/ZFY factors{2.3.3.65} | 99211 | 22764 | ZFX_MOUSE | P17012 | |
| ZIC1_MOUSE.H10MO.B |  |
Zic1 | 9 | B | KGGGWGSKv | HOCOMOCO v9 | 34 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 106683 | 22771 | ZIC1_MOUSE | P46684 | ||||
| ZIC2_MOUSE.H10MO.C |  |
Zic2 | 9 | C | dKGGdGbKv | HOCOMOCO v9 | 36 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 106679 | ZIC2_MOUSE | Q62520 | |||||
| ZIC3_MOUSE.H10MO.C |  |
Zic3 | 9 | C | bGGGKRvYb | HOCOMOCO v9 | 33 | More than 3 adjacent zinc finger factors{2.3.3} | GLI-like factors{2.3.3.1} | 106676 | 22773 | ZIC3_MOUSE | Q62521 | ||||
| ZN143_MOUSE.H10MO.D |  |
Znf143 | 22 | D | KMddKGMAKKMTGGGWRdKSbh | HOCOMOCO v9 | 26 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF76-like factors{2.3.3.28} | 1277969 | 20841 | ZN143_MOUSE | O70230 | ||||
| ZN148_MOUSE.H10MO.D |  |
Znf148 | 15 | D | dSvKbGGGGGhGGGR | HOCOMOCO v9 | 34 | More than 3 adjacent zinc finger factors{2.3.3} | ZNF148-like factors{2.3.3.13} | 1332234 | 22661 | ZN148_MOUSE | Q61624 | ||||
| ZN423_MOUSE.H10MO.D |  |
Znf423 | 15 | D | GGCACCCAAGGGTGC | HOCOMOCO v9 | 22 | Factors with multiple dispersed zinc fingers{2.3.4} | ZNF423-like factors{2.3.4.12} | 1891217 | 94187 | ZN423_MOUSE | Q80TS5 | ||||
| ENOA_MOUSE.H10MO.A RETRACTED!!! |  |
Eno1 | 13 | A | vdSCAYGTGbbhv | HOCOMOCO v9 | 1994 | 95393 | 13806; 433182 | ENOA_MOUSE | P17182 | Retracted motif! |