Dinucleotide PWMs for MOUSE transcription factors
| Model | LOGO | Transcription factor | Model length | Quality | Consensus | Origin | wAUC | Best AUC | Benchmark datasets | Aligned words | TF family | TF subfamily | MGI | EntrezGene | UniProt ID | UniProt AC | Comment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BCL6_MOUSE.H10DI.C |  |
Bcl6 | 16 | C | nndMYbTCYAGKRAhn | ChIP-Seq | 0.7353697537191268 | 0.7759557538066195 | 3 | 504 | More than 3 adjacent zinc finger factors{2.3.3} | BCL6 factors{2.3.3.22} | 107187 | 12053 | BCL6_MOUSE | P41183 | |
| CDX4_MOUSE.H10DI.A |  |
Cdx4 | 14 | A | nWRATTTAYKRbhn | ChIP-Seq | 0.8556649397174473 | 0.8716569321731612 | 2 | 513 | HOX-related factors{3.1.1} | CDX (Caudal type homeobox){3.1.1.9} | 88362 | 12592 | CDX4_MOUSE | Q07424 | |
| CEBPB_MOUSE.H10DI.A |  |
Cebpb | 21 | A | nnRTKRYGCAAYhnnnnnnnn | ChIP-Seq | 0.9303774581142188 | 0.9474797264071769 | 10 | 500 | C/EBP-related{1.1.8} | C/EBP{1.1.8.1} | 88373 | 12608 | CEBPB_MOUSE | P28033 | |
| COE1_MOUSE.H10DI.C |  |
Ebf1 | 23 | C | nvndbYCCCWKGGGRvhnnnnnn | ChIP-Seq | 0.9346269203953789 | 0.9346269203953789 | 1 | 506 | Early B-Cell Factor-related factors{6.1.5} | EBF1 (COE1){6.1.5.0.1} | 95275 | 13591 | COE1_MOUSE | Q07802 | |
| CRX_MOUSE.H10DI.A |  |
Crx | 18 | A | nnnnhdvddRGATTAdnn | ChIP-Seq | 0.862689719482612 | 0.8724564705431574 | 2 | 500 | Paired-related HD factors{3.1.3} | OTX{3.1.3.17} | 1194883 | 12951 | CRX_MOUSE | O54751 | |
| CTCF_MOUSE.H10DI.A |  |
Ctcf | 23 | A | nYbdCCAShAGRKGRCRbbvndb | ChIP-Seq | 0.9828675552675289 | 0.9954072691558203 | 35 | 502 | More than 3 adjacent zinc finger factors{2.3.3} | CTCF-like factors{2.3.3.50} | 109447 | 13018 | CTCF_MOUSE | Q61164 | |
| E2F4_MOUSE.H10DI.C |  |
E2f4 | 22 | C | vvvvddGGCGGGRvnnbvvvvn | ChIP-Seq | 0.7542100464699116 | 0.7924096002243018 | 4 | 499 | E2F-related factors{3.3.2} | E2F{3.3.2.1} | 103012 | 104394 | E2F4_MOUSE | Q8R0K9 | |
| ERR2_MOUSE.H10DI.C |  |
Esrrb | 23 | C | nndbnbSAAGGTCAnvbnvnvnn | ChIP-Seq | 0.9683998673745083 | 0.9683998673745083 | 1 | 529 | Steroid hormone receptors (NR3){2.1.1} | ER-like receptors (NR3A&B){2.1.1.2} | 1346832 | 26380 | ERR2_MOUSE | Q61539 | |
| EVX1_MOUSE.H10DI.C |  |
Evx1 | 17 | C | nWRATTKATKRbhhnnn | ChIP-Seq | 0.8342128522087124 | 0.8342128522087123 | 1 | 500 | HOX-related factors{3.1.1} | EVX (Even-skipped homolog){3.1.1.10} | 95461 | 14028 | EVX1_MOUSE | P23683 | |
| EVX2_MOUSE.H10DI.C |  |
Evx2 | 15 | C | nbbTTTATKRYhbbn | ChIP-Seq | 0.9311654294920892 | 0.9311654294920892 | 1 | 534 | HOX-related factors{3.1.1} | EVX (Even-skipped homolog){3.1.1.10} | 95462 | 14029 | EVX2_MOUSE | P49749 | |
| FLI1_MOUSE.H10DI.A |  |
Fli1 | 23 | A | nbnnnvvvRCMGGAAGYSvvdbn | ChIP-Seq | 0.8591646810780967 | 0.9329776164245106 | 3 | 457 | Ets-related factors{3.5.2} | Ets-like factors{3.5.2.1} | 95554 | 14247 | FLI1_MOUSE | P26323 | |
| FOXA2_MOUSE.H10DI.B |  |
Foxa2 | 20 | B | nnnbhnhTRTTTACWYWdnn | ChIP-Seq | 0.8643172053579138 | 0.9346536602102022 | 2 | 511 | Forkhead box (FOX) factors{3.3.1} | FOXA{3.3.1.1} | 1347476 | 15376 | FOXA2_MOUSE | P35583 | |
| GATA1_MOUSE.H10DI.A |  |
Gata1 | 21 | A | nnbnnnnnndbWGATAASvnn | ChIP-Seq | 0.8852083950439796 | 0.9304282739217927 | 13 | 515 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95661 | 14460 | GATA1_MOUSE | P17679 | |
| GATA2_MOUSE.H10DI.A |  |
Gata2 | 23 | A | nnnnnnnnnnnnvvWGATAASvn | ChIP-Seq | 0.850803462219922 | 0.9011483742840624 | 5 | 501 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95662 | 14461 | GATA2_MOUSE | O09100 | |
| GATA3_MOUSE.H10DI.A |  |
Gata3 | 23 | A | ndnddndbdnnvAGATAAvddnn | ChIP-Seq | 0.8287681066183508 | 0.8462938671120581 | 2 | 504 | GATA-type zinc fingers{2.2.1} | Two zinc-finger GATA factors{2.2.1.1} | 95663 | 14462 | GATA3_MOUSE | P23772 | |
| GFI1B_MOUSE.H10DI.C | 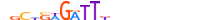 |
Gfi1b | 22 | C | nRYWSWGATTKvnnnnnnnnnn | ChIP-Seq | 0.825953473931337 | 0.825953473931337 | 1 | 525 | More than 3 adjacent zinc finger factors{2.3.3} | GFI1 factors{2.3.3.21} | 1276578 | 14582 | GFI1B_MOUSE | O70237 | |
| JUND_MOUSE.H10DI.C |  |
Jund | 19 | C | ndnKbhhvdWMTGASTMWn | ChIP-Seq | 0.8658295450293818 | 0.8658295450293818 | 1 | 501 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96648 | 16478 | JUND_MOUSE | P15066 | |
| JUN_MOUSE.H10DI.C |  |
Jun | 22 | C | nddKddvRvdRTGASWhMnhnn | ChIP-Seq | 0.860163854352609 | 0.8601638543526091 | 1 | 506 | Jun-related factors{1.1.1} | Jun factors{1.1.1.1} | 96646 | 16476 | JUN_MOUSE | P05627 | |
| KLF4_MOUSE.H10DI.C |  |
Klf4 | 22 | C | nvnnvvndGGGYGKGGYbbnvn | ChIP-Seq | 0.9287471424912743 | 0.9287471424912743 | 1 | 520 | Three-zinc finger Krüppel-related factors{2.3.1} | Krüppel-like factors{2.3.1.2} | 1342287 | 16600 | KLF4_MOUSE | Q60793 | |
| MAFK_MOUSE.H10DI.A |  |
Mafk | 23 | A | ndWhnTGCTGASTMAKCWnhhnn | ChIP-Seq | 0.9746003345724599 | 0.9761741452190391 | 2 | 437 | Maf-related factors{1.1.3} | Small Maf factors{1.1.3.2} | 99951 | 17135 | MAFK_MOUSE | Q61827 | |
| MAX_MOUSE.H10DI.A |  |
Max | 23 | A | nvvCACGTGbhbnbnbnnnnnbn | ChIP-Seq | 0.8306275995481456 | 0.8968438650315729 | 4 | 514 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 96921 | MAX_MOUSE | P28574 | ||
| MXI1_MOUSE.H10DI.B |  |
Mxi1 | 23 | B | nvvvCAYGTGSvnvvnbvvvnbn | ChIP-Seq | 0.7838391847178962 | 0.8008708723194874 | 2 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 97245 | 17859 | MXI1_MOUSE | P50540 | |
| MYB_MOUSE.H10DI.A |  |
Myb | 22 | A | nndvdKRCMGTTRvnnnnbnnn | ChIP-Seq | 0.807545787856477 | 0.8137724634568896 | 2 | 511 | Myb/SANT domain factors{3.5.1} | Myb-like factors{3.5.1.1} | 97249 | 17863 | MYB_MOUSE | P06876 | |
| MYC_MOUSE.H10DI.A |  |
Myc | 19 | A | nndvMCACGTGSnvvnnvn | ChIP-Seq | 0.8562701197561228 | 0.8929962114698462 | 3 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 97250 | 17869 | MYC_MOUSE | P01108 | |
| MYOD1_MOUSE.H10DI.C |  |
Myod1 | 23 | C | nnnvCAGCTGYYbbhbbbnnbbn | ChIP-Seq | 0.9352638874024874 | 0.9352638874024874 | 1 | 500 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97275 | 17927 | MYOD1_MOUSE | P10085 | |
| MYOG_MOUSE.H10DI.C |  |
Myog | 22 | C | nvvvvnRRCAGCTGbndnnnnn | ChIP-Seq | 0.9402627128393427 | 0.9402627128393428 | 1 | 461 | MyoD / ASC-related factors{1.2.2} | Myogenic transcription factors{1.2.2.1} | 97276 | 17928 | MYOG_MOUSE | P12979 | |
| NANOG_MOUSE.H10DI.A |  |
Nanog | 22 | A | nbbYWTTvWnWTKYWRAdndnn | ChIP-Seq | 0.7972551782413955 | 0.8613327589831379 | 4 | 513 | NK-related factors{3.1.2} | NANOG{3.1.2.12} | 1919200 | 71950 | NANOG_MOUSE | Q80Z64 | |
| NDF1_MOUSE.H10DI.C |  |
Neurod1 | 23 | C | bbvvvvnddRRMAGATGGYnhvv | ChIP-Seq | 0.9092513609210184 | 0.9092513609210184 | 1 | 731 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1339708 | 18012 | NDF1_MOUSE | Q60867 | |
| NFYA_MOUSE.H10DI.C |  |
Nfya | 23 | C | bdRMvvhhhhvRMMAATSRKARv | ChIP-Seq | 0.8070458887767631 | 0.8070458887767631 | 1 | 579 | Heteromeric CCAAT-binding factors{4.2.1} | NF-YA (CP1A, CBF-B){4.2.1.0.1} | 97316 | 18044 | NFYA_MOUSE | P23708 | |
| NR1D1_MOUSE.H10DI.D | 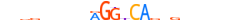 |
Nr1d1 | 23 | D | nvdddvvnnRGGbCAvdKdbnnn | ChIP-Seq | 0.7268982058707454 | 0.7268982058707455 | 1 | 513 | Thyroid hormone receptor-related factors (NR1){2.1.2} | Rev-ErbA (NR1D){2.1.2.3} | 2444210 | 217166 | NR1D1_MOUSE | Q3UV55 | |
| NR5A2_MOUSE.H10DI.A |  |
Nr5a2 | 16 | A | ndbYCAAGGYCRnnnn | ChIP-Seq | 0.9128812709810099 | 0.9197057894990996 | 2 | 501 | FTZ-F1-related receptors (NR5){2.1.5} | LRH-1 (NR5A2){2.1.5.0.2} | 1346834 | 26424 | NR5A2_MOUSE | P45448 | |
| OLIG2_MOUSE.H10DI.B |  |
Olig2 | 23 | B | nMCAKCTGbbnnShdbvhbbbnn | ChIP-Seq | 0.8010924228351114 | 0.8146213135592371 | 2 | 502 | Tal-related factors{1.2.3} | Neurogenin / Atonal-like factors{1.2.3.4} | 1355331 | 50913 | OLIG2_MOUSE | Q9EQW6 | |
| PO5F1_MOUSE.H10DI.A | 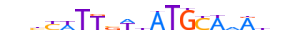 |
Pou5f1 | 19 | A | nbbYWTTvWhATGCAdRdn | ChIP-Seq | 0.902146413845935 | 0.9725630727094943 | 6 | 505 | POU domain factors{3.1.10} | POU5 (Oct-3/4-like factors){3.1.10.5} | 101893 | 18999 | PO5F1_MOUSE | P20263 | |
| PPARG_MOUSE.H10DI.A |  |
Pparg | 23 | A | nddvnhWbWRKGbbARAGGKbWn | ChIP-Seq | 0.8358730078031252 | 0.895069341589453 | 6 | 503 | Thyroid hormone receptor-related factors (NR1){2.1.2} | PPAR (NR1C){2.1.2.5} | 97747 | 19016 | PPARG_MOUSE | P37238 | |
| PRD14_MOUSE.H10DI.C |  |
Prdm14 | 14 | C | ndSWTAGRGMMShn | ChIP-Seq | 0.8016261451947589 | 0.8016261451947589 | 1 | 511 | More than 3 adjacent zinc finger factors{2.3.3} | unclassified{2.3.3.0} | 3588194 | 383491 | PRD14_MOUSE | E9Q3T6 | |
| REST_MOUSE.H10DI.A |  |
Rest | 23 | A | bYYWSSRYSdbGGWCAGYKbYhn | ChIP-Seq | 0.9141971050889611 | 0.9404404842238376 | 2 | 532 | Factors with multiple dispersed zinc fingers{2.3.4} | unclassified{2.3.4.0} | 104897 | 19712 | REST_MOUSE | Q8VIG1 | |
| RFX1_MOUSE.H10DI.C |  |
Rfx1 | 23 | C | bbbbSbGTTGCCWdGGvvMSvvb | ChIP-Seq | 0.9000134299663253 | 0.9000134299663254 | 1 | 440 | RFX-related factors{3.3.3} | RFX1 (EF-C){3.3.3.0.1} | 105982 | 19724 | RFX1_MOUSE | P48377 | |
| RXRA_MOUSE.H10DI.B |  |
Rxra | 23 | B | nvdvRGKKSARGGbSWbbbhnvd | ChIP-Seq | 0.7654882691921001 | 0.8069879887626238 | 3 | 507 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98214 | 20181 | RXRA_MOUSE | P28700 | |
| RXRG_MOUSE.H10DI.C |  |
Rxrg | 23 | C | vddvdbbvdRAGKTCARGGhYRb | ChIP-Seq | 0.8616538648421981 | 0.8616538648421981 | 1 | 520 | RXR-related receptors (NR2){2.1.3} | Retinoid X receptors (NR2B){2.1.3.1} | 98216 | 20183 | RXRG_MOUSE | P28705 | |
| SPI1_MOUSE.H10DI.A |  |
Spi1 | 23 | A | nvvWRWKRGGAARTGvvnnnnnn | ChIP-Seq | 0.9587865003775525 | 0.9891271635038471 | 7 | 500 | Ets-related factors{3.5.2} | Spi-like factors{3.5.2.5} | 98282 | 20375 | SPI1_MOUSE | P17433 | |
| STA5A_MOUSE.H10DI.A |  |
Stat5a | 14 | A | ndnTTCYTRGAAnn | ChIP-Seq | 0.9185652920874283 | 0.961199934260125 | 6 | 500 | STAT factors{6.2.1} | STAT5A{6.2.1.0.5} | 103036 | 20850 | STA5A_MOUSE | P42230 | |
| STA5B_MOUSE.H10DI.C |  |
Stat5b | 14 | C | ndTTCYTRGAAnhn | ChIP-Seq | 0.9507193214364849 | 0.9507193214364849 | 1 | 503 | STAT factors{6.2.1} | STAT5B{6.2.1.0.6} | 103035 | 20851 | STA5B_MOUSE | P42232 | |
| STAT1_MOUSE.H10DI.A | 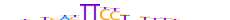 |
Stat1 | 23 | A | nbhnYhWYTTCCYbYhbhhhhdn | ChIP-Seq | 0.7906657931512691 | 0.818755033918522 | 6 | 502 | STAT factors{6.2.1} | STAT1{6.2.1.0.1} | 103063 | STAT1_MOUSE | P42225 | ||
| STAT3_MOUSE.H10DI.A |  |
Stat3 | 18 | A | nvnbTKMYdGGAAvbvvn | ChIP-Seq | 0.7798264758952239 | 0.814932416378456 | 4 | 436 | STAT factors{6.2.1} | STAT3{6.2.1.0.3} | 103038 | 20848 | STAT3_MOUSE | P42227 | |
| STAT4_MOUSE.H10DI.A |  |
Stat4 | 22 | A | nvnnnbbnbTTCCYdKMAdnnn | ChIP-Seq | 0.8592520347735834 | 0.8744564302186991 | 2 | 502 | STAT factors{6.2.1} | STAT4{6.2.1.0.4} | 103062 | 20849 | STAT4_MOUSE | P42228 | |
| STAT6_MOUSE.H10DI.C | 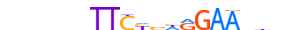 |
Stat6 | 20 | C | nnnnnbTTCYbhRGAAvhnn | ChIP-Seq | 0.8262492362491791 | 0.8262492362491792 | 1 | 500 | STAT factors{6.2.1} | STAT6{6.2.1.0.7} | 103034 | 20852 | STAT6_MOUSE | P52633 | |
| TAL1_MOUSE.H10DI.A |  |
Tal1 | 23 | A | nnnbbKbnnnnndvWGATAARvn | ChIP-Seq | 0.920395509856274 | 0.973788791908167 | 11 | 502 | Tal-related factors{1.2.3} | Tal / HEN-like factors{1.2.3.1} | 98480 | 21349 | TAL1_MOUSE | P22091 | |
| TBX20_MOUSE.H10DI.C |  |
Tbx20 | 21 | C | nnnnnnnvdSTGnTGACAKvn | ChIP-Seq | 0.8141096795432899 | 0.8141096795432899 | 1 | 502 | TBX1-related factors{6.5.3} | TBX20{6.5.3.0.5} | 1888496 | 57246 | TBX20_MOUSE | Q9ES03 | |
| TF2L1_MOUSE.H10DI.C |  |
Tfcp2l1 | 23 | C | nndRdCYRGhhdRARCYRGhhhn | ChIP-Seq | 0.9573716300086532 | 0.9573716300086532 | 1 | 488 | CP2-related factors{6.7.2} | CP2-L1 (LBP-9, CRTR-1){6.7.2.0.2} | 2444691 | 81879 | TF2L1_MOUSE | Q3UNW5 | |
| TF65_MOUSE.H10DI.C |  |
Rela | 23 | C | nRGRRWKYCCMndvYKdbnhMMn | ChIP-Seq | 0.8395047729499199 | 0.8395047729499199 | 1 | 500 | NF-kappaB-related factors{6.1.1} | NF-kappaB p65 subunit-like factors{6.1.1.2} | 103290 | 19697 | TF65_MOUSE | Q04207 | |
| TFE2_MOUSE.H10DI.A |  |
Tcf3 | 23 | A | ndSCAGSTGbbvbdSbRKvbdbb | ChIP-Seq | 0.8599612631436858 | 0.8709568721113843 | 3 | 584 | E2A-related factors{1.2.1} | E2A (TCF-3, ITF-1){1.2.1.0.1} | 98510 | 21423 | TFE2_MOUSE | P15806 | |
| USF2_MOUSE.H10DI.A |  |
Usf2 | 23 | A | ndGTCACGTGvbbnvnnbnvnbb | ChIP-Seq | 0.9803407420232653 | 0.9899993048469995 | 2 | 517 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 99961 | 22282 | USF2_MOUSE | Q64705 |