Model info
| Transcription factor | Spib | ||||||||
| Model | SPIB_MOUSE.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 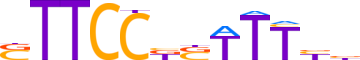 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 12 | ||||||||
| Quality | B | ||||||||
| Consensus | vvAAASMGGAAS | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 82 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Spi-like factors{3.5.2.5} | ||||||||
| MGI | 892986 | ||||||||
| EntrezGene | 272382 | ||||||||
| UniProt ID | SPIB_MOUSE | ||||||||
| UniProt AC | O35906 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 38.048 | 16.033 | 17.075 | 10.963 |
| 02 | 43.006 | 9.028 | 24.08 | 6.004 |
| 03 | 70.11 | 0.0 | 6.004 | 6.004 |
| 04 | 76.114 | 0.0 | 0.0 | 6.004 |
| 05 | 59.017 | 0.0 | 0.0 | 23.102 |
| 06 | 10.007 | 27.104 | 43.006 | 2.001 |
| 07 | 41.158 | 24.994 | 13.965 | 2.001 |
| 08 | 4.936 | 0.0 | 77.182 | 0.0 |
| 09 | 1.023 | 0.0 | 81.095 | 0.0 |
| 10 | 82.118 | 0.0 | 0.0 | 0.0 |
| 11 | 82.118 | 0.0 | 0.0 | 0.0 |
| 12 | 5.304 | 26.315 | 47.242 | 3.258 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.593 | -0.233 | -0.174 | -0.584 |
| 02 | 0.712 | -0.759 | 0.152 | -1.113 |
| 03 | 1.192 | -2.977 | -1.113 | -1.113 |
| 04 | 1.272 | -2.977 | -2.977 | -1.113 |
| 05 | 1.022 | -2.977 | -2.977 | 0.112 |
| 06 | -0.666 | 0.265 | 0.712 | -1.942 |
| 07 | 0.67 | 0.188 | -0.362 | -1.942 |
| 08 | -1.276 | -2.977 | 1.286 | -2.977 |
| 09 | -2.32 | -2.977 | 1.335 | -2.977 |
| 10 | 1.347 | -2.977 | -2.977 | -2.977 |
| 11 | 1.347 | -2.977 | -2.977 | -2.977 |
| 12 | -1.217 | 0.237 | 0.804 | -1.602 |