Model info
| Transcription factor | Atoh1 | ||||||||
| Model | ATOH1_MOUSE.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 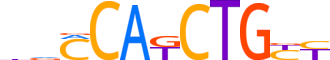 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 11 | ||||||||
| Quality | C | ||||||||
| Consensus | RRCAGMTGKbv | ||||||||
| wAUC | 0.8927500766335188 | ||||||||
| Best AUC | 0.8927500766335188 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | Tal-related factors{1.2.3} | ||||||||
| TF subfamily | Neurogenin / Atonal-like factors{1.2.3.4} | ||||||||
| MGI | 104654 | ||||||||
| EntrezGene | 11921 | ||||||||
| UniProt ID | ATOH1_MOUSE | ||||||||
| UniProt AC | P48985 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 271.88 | 13.701 | 195.351 | 19.069 |
| 02 | 148.55 | 67.467 | 283.036 | 0.947 |
| 03 | 4.779 | 495.221 | 0.0 | 0.0 |
| 04 | 500.0 | 0.0 | 0.0 | 0.0 |
| 05 | 2.005 | 0.0 | 493.003 | 4.991 |
| 06 | 262.333 | 221.848 | 15.819 | 0.0 |
| 07 | 7.795 | 2.008 | 0.0 | 490.197 |
| 08 | 0.956 | 0.0 | 498.07 | 0.974 |
| 09 | 7.938 | 34.659 | 378.192 | 79.21 |
| 10 | 56.837 | 221.198 | 85.564 | 136.402 |
| 11 | 198.208 | 105.823 | 124.216 | 71.753 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.77 | -2.116 | 0.442 | -1.814 |
| 02 | 0.171 | -0.606 | 0.81 | -3.924 |
| 03 | -2.995 | 1.367 | -4.4 | -4.4 |
| 04 | 1.377 | -4.4 | -4.4 | -4.4 |
| 05 | -3.571 | -4.4 | 1.363 | -2.962 |
| 06 | 0.735 | 0.568 | -1.986 | -4.4 |
| 07 | -2.605 | -3.57 | -4.4 | 1.357 |
| 08 | -3.92 | -4.4 | 1.373 | -3.913 |
| 09 | -2.59 | -1.251 | 1.099 | -0.449 |
| 10 | -0.774 | 0.565 | -0.373 | 0.086 |
| 11 | 0.456 | -0.164 | -0.006 | -0.546 |