Model info
| Transcription factor | Nr1i3 | ||||||||
| Model | NR1I3_MOUSE.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 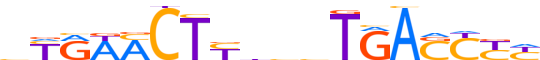 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Consensus | RRGSTCAvvvRAGKWYRd | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 28 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | Vitamin D receptor (NR1I){2.1.2.4} | ||||||||
| MGI | 1346307 | ||||||||
| EntrezGene | 12355 | ||||||||
| UniProt ID | NR1I3_MOUSE | ||||||||
| UniProt AC | O35627 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 9.049 | 0.0 | 13.84 | 5.11 |
| 02 | 12.776 | 2.129 | 13.095 | 0.0 |
| 03 | 5.004 | 0.0 | 20.973 | 2.023 |
| 04 | 0.0 | 3.939 | 20.335 | 3.726 |
| 05 | 0.0 | 0.0 | 0.958 | 27.042 |
| 06 | 0.0 | 22.996 | 1.916 | 3.087 |
| 07 | 25.977 | 2.023 | 0.0 | 0.0 |
| 08 | 8.943 | 4.578 | 10.008 | 4.471 |
| 09 | 7.133 | 5.43 | 10.859 | 4.578 |
| 10 | 11.179 | 5.536 | 7.452 | 3.833 |
| 11 | 16.289 | 2.023 | 8.73 | 0.958 |
| 12 | 25.338 | 0.0 | 2.662 | 0.0 |
| 13 | 1.065 | 0.0 | 26.935 | 0.0 |
| 14 | 0.0 | 1.81 | 5.962 | 20.228 |
| 15 | 4.897 | 2.981 | 0.0 | 20.122 |
| 16 | 0.0 | 19.802 | 2.023 | 6.175 |
| 17 | 19.909 | 2.023 | 3.939 | 2.129 |
| 18 | 5.137 | 3.966 | 10.034 | 8.863 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.232 | -2.241 | 0.628 | -0.276 |
| 02 | 0.552 | -0.972 | 0.576 | -2.241 |
| 03 | -0.294 | -2.241 | 1.024 | -1.009 |
| 04 | -2.241 | -0.496 | 0.994 | -0.541 |
| 05 | -2.241 | -2.241 | -1.475 | 1.269 |
| 06 | -2.241 | 1.113 | -1.047 | -0.692 |
| 07 | 1.23 | -1.009 | -2.241 | -2.241 |
| 08 | 0.222 | -0.37 | 0.325 | -0.39 |
| 09 | 0.017 | -0.224 | 0.401 | -0.37 |
| 10 | 0.428 | -0.207 | 0.056 | -0.518 |
| 11 | 0.782 | -1.009 | 0.2 | -1.475 |
| 12 | 1.206 | -2.241 | -0.807 | -2.241 |
| 13 | -1.418 | -2.241 | 1.266 | -2.241 |
| 14 | -2.241 | -1.086 | -0.142 | 0.989 |
| 15 | -0.313 | -0.72 | -2.241 | 0.984 |
| 16 | -2.241 | 0.969 | -1.009 | -0.111 |
| 17 | 0.974 | -1.009 | -0.496 | -0.972 |
| 18 | -0.272 | -0.49 | 0.327 | 0.213 |