Model info
| Transcription factor | Nr3c1 | ||||||||
| Model | GCR_MOUSE.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 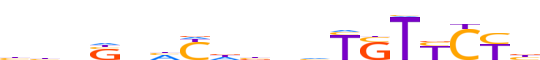 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Consensus | vRGRASAdnvhRdnSndd | ||||||||
| wAUC | 0.7467282195273168 | ||||||||
| Best AUC | 0.757625051561599 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 4198 | ||||||||
| TF family | Steroid hormone receptors (NR3){2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C){2.1.1.1} | ||||||||
| MGI | 95824 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | GCR_MOUSE | ||||||||
| UniProt AC | P06537 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1129.592 | 904.237 | 1807.363 | 281.005 |
| 02 | 2413.167 | 43.725 | 1641.054 | 24.25 |
| 03 | 521.354 | 80.966 | 3467.337 | 52.539 |
| 04 | 2816.467 | 284.186 | 822.166 | 199.377 |
| 05 | 3847.698 | 50.096 | 111.135 | 113.267 |
| 06 | 240.929 | 2963.207 | 832.499 | 85.56 |
| 07 | 3338.714 | 113.992 | 367.264 | 302.226 |
| 08 | 791.544 | 485.085 | 1594.931 | 1250.635 |
| 09 | 1520.14 | 779.229 | 801.152 | 1021.675 |
| 10 | 1224.342 | 1299.994 | 1230.157 | 367.702 |
| 11 | 1299.273 | 580.13 | 366.139 | 1876.653 |
| 12 | 461.225 | 246.014 | 3070.114 | 344.843 |
| 13 | 588.589 | 345.953 | 957.889 | 2229.765 |
| 14 | 813.465 | 1213.193 | 906.934 | 1188.604 |
| 15 | 408.033 | 2749.102 | 559.13 | 405.931 |
| 16 | 1057.156 | 1403.754 | 507.771 | 1153.515 |
| 17 | 1371.743 | 427.692 | 1375.528 | 947.233 |
| 18 | 1130.47 | 566.318 | 1699.453 | 725.955 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.092 | -0.13 | 0.561 | -1.294 |
| 02 | 0.85 | -3.115 | 0.464 | -3.669 |
| 03 | -0.679 | -2.52 | 1.212 | -2.939 |
| 04 | 1.004 | -1.283 | -0.225 | -1.634 |
| 05 | 1.316 | -2.985 | -2.211 | -2.192 |
| 06 | -1.447 | 1.055 | -0.213 | -2.467 |
| 07 | 1.174 | -2.186 | -1.028 | -1.222 |
| 08 | -0.263 | -0.751 | 0.436 | 0.193 |
| 09 | 0.388 | -0.279 | -0.251 | -0.009 |
| 10 | 0.172 | 0.232 | 0.177 | -1.027 |
| 11 | 0.231 | -0.573 | -1.031 | 0.598 |
| 12 | -0.801 | -1.426 | 1.09 | -1.091 |
| 13 | -0.559 | -1.088 | -0.073 | 0.771 |
| 14 | -0.236 | 0.163 | -0.128 | 0.142 |
| 15 | -0.923 | 0.98 | -0.61 | -0.929 |
| 16 | 0.025 | 0.309 | -0.706 | 0.113 |
| 17 | 0.285 | -0.877 | 0.288 | -0.084 |
| 18 | 0.092 | -0.597 | 0.499 | -0.35 |