Model info
| Transcription factor | Rxra | ||||||||
| Model | RXRA_MOUSE.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 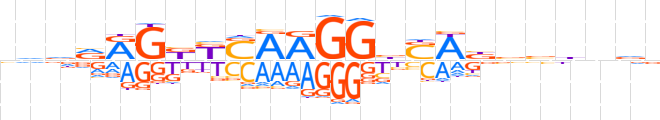 | ||||||||
| LOGO (reverse complement) | 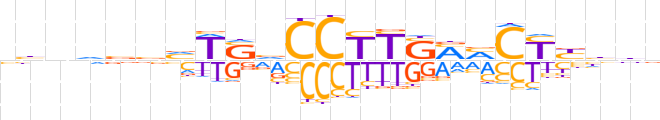 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | B | ||||||||
| Consensus | nvdvRGKKSARGGbSWbbbhnvd | ||||||||
| wAUC | 0.7654882691921001 | ||||||||
| Best AUC | 0.8069879887626238 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 507 | ||||||||
| TF family | RXR-related receptors (NR2){2.1.3} | ||||||||
| TF subfamily | Retinoid X receptors (NR2B){2.1.3.1} | ||||||||
| MGI | 98214 | ||||||||
| EntrezGene | 20181 | ||||||||
| UniProt ID | RXRA_MOUSE | ||||||||
| UniProt AC | P28700 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.692 | 12.578 | 53.201 | 4.915 | 115.635 | 39.287 | 12.369 | 26.52 | 25.846 | 27.968 | 47.806 | 4.105 | 12.498 | 43.967 | 20.611 | 23.95 |
| 02 | 57.958 | 21.878 | 91.244 | 10.59 | 65.646 | 16.855 | 6.854 | 34.446 | 28.706 | 17.173 | 59.177 | 28.931 | 10.322 | 4.526 | 20.476 | 24.165 |
| 03 | 54.532 | 14.231 | 87.345 | 6.524 | 32.838 | 10.903 | 8.321 | 8.37 | 59.478 | 11.764 | 95.969 | 10.539 | 16.962 | 12.939 | 62.362 | 5.869 |
| 04 | 108.11 | 0.0 | 48.145 | 7.556 | 29.337 | 0.0 | 5.578 | 14.921 | 188.018 | 1.962 | 51.551 | 12.466 | 15.881 | 0.949 | 10.194 | 4.279 |
| 05 | 7.371 | 8.816 | 309.88 | 15.28 | 0.0 | 0.0 | 0.0 | 2.911 | 6.62 | 1.015 | 92.982 | 14.85 | 2.521 | 3.32 | 25.936 | 7.446 |
| 06 | 1.91 | 2.941 | 7.949 | 3.712 | 0.0 | 5.929 | 0.0 | 7.222 | 27.885 | 28.083 | 145.447 | 227.382 | 1.078 | 6.387 | 9.615 | 23.407 |
| 07 | 6.365 | 3.771 | 13.342 | 7.396 | 8.58 | 13.58 | 2.497 | 18.683 | 12.75 | 19.817 | 34.557 | 95.887 | 6.653 | 21.259 | 21.412 | 212.399 |
| 08 | 8.202 | 13.455 | 10.905 | 1.786 | 15.03 | 38.063 | 1.74 | 3.594 | 4.518 | 26.582 | 36.037 | 4.67 | 11.763 | 289.299 | 20.42 | 12.883 |
| 09 | 25.393 | 0.0 | 11.984 | 2.135 | 336.656 | 18.446 | 5.558 | 6.741 | 31.231 | 16.747 | 18.398 | 2.726 | 11.563 | 3.699 | 5.828 | 1.843 |
| 10 | 315.755 | 6.131 | 73.396 | 9.56 | 30.226 | 0.961 | 4.194 | 3.51 | 24.232 | 0.879 | 14.555 | 2.102 | 5.865 | 1.101 | 5.53 | 0.949 |
| 11 | 17.35 | 1.07 | 353.98 | 3.679 | 4.084 | 2.005 | 1.84 | 1.143 | 5.472 | 0.0 | 88.192 | 4.011 | 0.0 | 0.0 | 14.319 | 1.802 |
| 12 | 2.107 | 1.803 | 21.072 | 1.924 | 0.961 | 0.0 | 2.114 | 0.0 | 28.877 | 4.933 | 418.172 | 6.35 | 1.74 | 0.0 | 6.057 | 2.837 |
| 13 | 5.274 | 11.754 | 14.647 | 2.01 | 1.94 | 3.902 | 0.0 | 0.894 | 45.581 | 97.47 | 87.917 | 216.448 | 0.923 | 2.651 | 1.691 | 5.846 |
| 14 | 7.393 | 28.168 | 14.292 | 3.865 | 6.689 | 87.226 | 6.574 | 15.287 | 10.834 | 51.632 | 22.839 | 18.948 | 2.718 | 181.664 | 21.002 | 19.815 |
| 15 | 18.142 | 2.351 | 1.671 | 5.471 | 303.992 | 10.533 | 10.043 | 24.123 | 42.224 | 4.193 | 11.022 | 7.268 | 28.292 | 4.987 | 13.394 | 11.242 |
| 16 | 37.693 | 61.099 | 207.64 | 86.218 | 4.346 | 8.83 | 4.969 | 3.918 | 4.562 | 2.961 | 21.752 | 6.855 | 0.961 | 6.608 | 27.995 | 12.54 |
| 17 | 7.81 | 17.15 | 21.679 | 0.923 | 22.373 | 35.113 | 6.397 | 15.615 | 40.738 | 85.144 | 99.78 | 36.695 | 6.55 | 48.721 | 29.63 | 24.63 |
| 18 | 22.805 | 17.174 | 25.287 | 12.205 | 18.887 | 120.047 | 11.429 | 35.764 | 21.24 | 57.117 | 52.99 | 26.138 | 6.146 | 40.443 | 15.147 | 16.127 |
| 19 | 24.77 | 17.46 | 13.267 | 13.581 | 49.19 | 25.243 | 7.054 | 153.294 | 19.099 | 20.99 | 31.203 | 33.561 | 8.14 | 26.088 | 24.005 | 32.002 |
| 20 | 35.248 | 7.248 | 49.505 | 9.198 | 44.8 | 9.687 | 4.682 | 30.612 | 22.728 | 8.074 | 21.517 | 23.21 | 36.955 | 58.748 | 88.155 | 48.58 |
| 21 | 36.489 | 18.626 | 73.269 | 11.347 | 50.362 | 15.76 | 3.643 | 13.993 | 51.912 | 24.12 | 71.955 | 15.872 | 8.399 | 16.766 | 56.19 | 30.245 |
| 22 | 30.171 | 14.749 | 86.766 | 15.476 | 29.49 | 13.818 | 7.205 | 24.759 | 35.499 | 29.028 | 104.248 | 36.281 | 16.692 | 21.188 | 21.526 | 12.05 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.117 | -0.89 | 0.529 | -1.784 | 1.302 | 0.228 | -0.906 | -0.16 | -0.185 | -0.107 | 0.423 | -1.95 | -0.896 | 0.34 | -0.408 | -0.26 |
| 02 | 0.614 | -0.349 | 1.066 | -1.056 | 0.738 | -0.605 | -1.472 | 0.098 | -0.082 | -0.587 | 0.635 | -0.074 | -1.081 | -1.86 | -0.414 | -0.251 |
| 03 | 0.554 | -0.77 | 1.022 | -1.519 | 0.051 | -1.028 | -1.288 | -1.282 | 0.64 | -0.955 | 1.116 | -1.061 | -0.599 | -0.862 | 0.687 | -1.618 |
| 04 | 1.234 | -4.398 | 0.43 | -1.38 | -0.06 | -4.398 | -1.666 | -0.724 | 1.786 | -2.598 | 0.498 | -0.899 | -0.663 | -3.162 | -1.093 | -1.912 |
| 05 | -1.403 | -1.233 | 2.285 | -0.701 | -4.398 | -4.398 | -4.398 | -2.259 | -1.505 | -3.113 | 1.084 | -0.728 | -2.384 | -2.142 | -0.182 | -1.394 |
| 06 | -2.62 | -2.25 | -1.332 | -2.041 | -4.398 | -1.609 | -4.398 | -1.423 | -0.11 | -0.103 | 1.53 | 1.976 | -3.069 | -1.539 | -1.149 | -0.283 |
| 07 | -1.542 | -2.027 | -0.833 | -1.4 | -1.259 | -0.815 | -2.393 | -0.504 | -0.877 | -0.446 | 0.102 | 1.115 | -1.5 | -0.377 | -0.37 | 1.908 |
| 08 | -1.302 | -0.824 | -1.028 | -2.676 | -0.717 | 0.197 | -2.697 | -2.07 | -1.862 | -0.158 | 0.143 | -1.831 | -0.955 | 2.217 | -0.417 | -0.867 |
| 09 | -0.203 | -4.398 | -0.937 | -2.527 | 2.368 | -0.517 | -1.67 | -1.488 | 0.001 | -0.611 | -0.519 | -2.316 | -0.971 | -2.044 | -1.625 | -2.65 |
| 10 | 2.304 | -1.577 | 0.849 | -1.155 | -0.031 | -3.153 | -1.93 | -2.092 | -0.249 | -3.215 | -0.748 | -2.54 | -1.619 | -3.054 | -1.674 | -3.162 |
| 11 | -0.577 | -3.075 | 2.418 | -2.049 | -1.954 | -2.58 | -2.651 | -3.026 | -1.684 | -4.398 | 1.032 | -1.971 | -4.398 | -4.398 | -0.764 | -2.668 |
| 12 | -2.538 | -2.668 | -0.386 | -2.614 | -3.153 | -4.398 | -2.535 | -4.398 | -0.076 | -1.781 | 2.585 | -1.545 | -2.697 | -4.398 | -1.589 | -2.281 |
| 13 | -1.718 | -0.956 | -0.742 | -2.577 | -2.607 | -1.996 | -4.398 | -3.203 | 0.376 | 1.131 | 1.029 | 1.927 | -3.181 | -2.341 | -2.72 | -1.622 |
| 14 | -1.401 | -0.1 | -0.766 | -2.005 | -1.495 | 1.021 | -1.512 | -0.7 | -1.034 | 0.499 | -0.307 | -0.49 | -2.319 | 1.752 | -0.389 | -0.446 |
| 15 | -0.533 | -2.445 | -2.73 | -1.684 | 2.266 | -1.062 | -1.107 | -0.253 | 0.3 | -1.93 | -1.018 | -1.417 | -0.096 | -1.77 | -0.829 | -0.999 |
| 16 | 0.187 | 0.667 | 1.885 | 1.009 | -1.897 | -1.231 | -1.774 | -1.992 | -1.853 | -2.244 | -0.355 | -1.472 | -3.153 | -1.507 | -0.106 | -0.893 |
| 17 | -1.348 | -0.588 | -0.358 | -3.181 | -0.327 | 0.117 | -1.537 | -0.679 | 0.264 | 0.997 | 1.155 | 0.161 | -1.515 | 0.442 | -0.05 | -0.233 |
| 18 | -0.308 | -0.587 | -0.207 | -0.919 | -0.493 | 1.339 | -0.983 | 0.135 | -0.378 | 0.6 | 0.525 | -0.174 | -1.575 | 0.257 | -0.709 | -0.648 |
| 19 | -0.227 | -0.57 | -0.838 | -0.815 | 0.451 | -0.208 | -1.445 | 1.583 | -0.483 | -0.39 | 0.001 | 0.073 | -1.309 | -0.176 | -0.258 | 0.026 |
| 20 | 0.121 | -1.419 | 0.458 | -1.192 | 0.359 | -1.142 | -1.829 | -0.018 | -0.312 | -1.317 | -0.366 | -0.291 | 0.168 | 0.628 | 1.031 | 0.439 |
| 21 | 0.155 | -0.507 | 0.847 | -0.99 | 0.475 | -0.67 | -2.058 | -0.786 | 0.505 | -0.253 | 0.829 | -0.664 | -1.279 | -0.61 | 0.583 | -0.03 |
| 22 | -0.033 | -0.735 | 1.015 | -0.688 | -0.055 | -0.799 | -1.425 | -0.228 | 0.128 | -0.071 | 1.198 | 0.15 | -0.614 | -0.381 | -0.365 | -0.932 |