Model info
| Transcription factor | Stat4 | ||||||||
| Model | STAT4_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 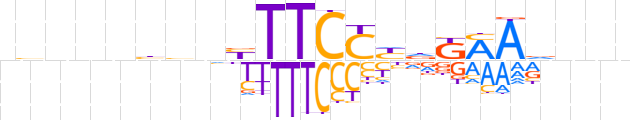 | ||||||||
| LOGO (reverse complement) | 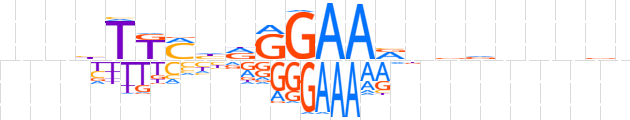 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Consensus | nvnnnbbnbTTCCYdKMAdnnn | ||||||||
| wAUC | 0.8592520347735834 | ||||||||
| Best AUC | 0.8744564302186991 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT4{6.2.1.0.4} | ||||||||
| MGI | 103062 | ||||||||
| EntrezGene | 20849 | ||||||||
| UniProt ID | STAT4_MOUSE | ||||||||
| UniProt AC | P42228 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 34.314 | 40.524 | 23.185 | 4.68 | 30.668 | 52.618 | 10.11 | 34.048 | 19.841 | 60.297 | 35.6 | 29.002 | 10.712 | 50.145 | 46.072 | 18.186 |
| 02 | 21.811 | 27.901 | 29.578 | 16.245 | 55.224 | 97.836 | 12.674 | 37.849 | 28.853 | 30.061 | 36.692 | 19.36 | 18.002 | 22.227 | 22.069 | 23.618 |
| 03 | 27.312 | 32.753 | 47.724 | 16.101 | 33.734 | 63.39 | 15.933 | 64.968 | 26.503 | 25.874 | 23.215 | 25.421 | 9.829 | 34.111 | 29.44 | 23.693 |
| 04 | 17.088 | 43.857 | 29.082 | 7.35 | 57.701 | 41.692 | 22.793 | 33.942 | 13.338 | 44.256 | 30.32 | 28.398 | 18.898 | 33.156 | 45.029 | 33.101 |
| 05 | 13.805 | 49.943 | 26.772 | 16.504 | 23.232 | 79.679 | 4.035 | 56.017 | 18.139 | 33.31 | 34.704 | 41.071 | 1.748 | 36.971 | 23.091 | 40.98 |
| 06 | 0.0 | 20.293 | 26.727 | 9.903 | 47.408 | 77.552 | 19.425 | 55.517 | 18.3 | 32.168 | 24.886 | 13.247 | 8.363 | 65.501 | 53.584 | 27.123 |
| 07 | 13.782 | 16.297 | 34.604 | 9.389 | 101.153 | 55.201 | 9.364 | 29.798 | 29.79 | 26.064 | 42.228 | 26.54 | 17.92 | 38.827 | 35.724 | 13.32 |
| 08 | 8.648 | 31.341 | 32.645 | 90.012 | 8.519 | 47.086 | 4.055 | 76.729 | 15.59 | 32.267 | 9.892 | 64.17 | 4.204 | 20.127 | 5.519 | 49.195 |
| 09 | 0.0 | 0.0 | 0.867 | 36.095 | 0.843 | 2.832 | 0.0 | 127.146 | 1.222 | 0.0 | 0.856 | 50.034 | 0.0 | 1.91 | 0.0 | 278.196 |
| 10 | 0.0 | 0.0 | 0.0 | 2.065 | 0.987 | 0.0 | 0.0 | 3.756 | 0.0 | 0.0 | 0.0 | 1.723 | 2.827 | 2.992 | 0.0 | 485.651 |
| 11 | 0.936 | 2.879 | 0.0 | 0.0 | 1.271 | 1.721 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.765 | 467.725 | 1.279 | 21.425 |
| 12 | 0.0 | 3.163 | 0.0 | 1.809 | 3.013 | 347.814 | 0.0 | 121.496 | 0.0 | 1.279 | 0.0 | 0.0 | 0.0 | 19.573 | 0.0 | 1.852 |
| 13 | 0.0 | 1.056 | 0.0 | 1.957 | 47.92 | 164.343 | 7.876 | 151.69 | 0.0 | 0.0 | 0.0 | 0.0 | 4.064 | 63.582 | 10.823 | 46.689 |
| 14 | 8.409 | 1.215 | 37.683 | 4.677 | 107.136 | 30.443 | 57.634 | 33.768 | 3.762 | 1.043 | 2.193 | 11.702 | 32.533 | 11.441 | 122.451 | 33.912 |
| 15 | 0.905 | 9.891 | 127.499 | 13.544 | 6.844 | 7.605 | 12.862 | 16.831 | 11.633 | 5.086 | 144.068 | 59.174 | 0.0 | 3.973 | 62.076 | 18.011 |
| 16 | 14.489 | 3.963 | 0.93 | 0.0 | 11.549 | 15.007 | 0.0 | 0.0 | 256.686 | 72.008 | 2.033 | 15.778 | 71.594 | 25.041 | 4.098 | 6.827 |
| 17 | 327.606 | 2.923 | 12.06 | 11.729 | 98.975 | 0.0 | 1.813 | 15.23 | 7.06 | 0.0 | 0.0 | 0.0 | 15.843 | 0.931 | 4.034 | 1.796 |
| 18 | 177.471 | 60.107 | 154.426 | 57.48 | 0.957 | 0.0 | 0.0 | 2.897 | 9.882 | 7.058 | 0.967 | 0.0 | 2.009 | 3.787 | 9.073 | 13.887 |
| 19 | 40.438 | 53.067 | 54.246 | 42.569 | 10.59 | 22.637 | 4.851 | 32.874 | 32.128 | 57.676 | 31.036 | 43.626 | 19.135 | 25.388 | 15.089 | 14.652 |
| 20 | 20.734 | 31.394 | 27.446 | 22.718 | 33.22 | 71.577 | 8.459 | 45.511 | 11.972 | 29.924 | 31.519 | 31.807 | 14.119 | 33.243 | 51.363 | 34.995 |
| 21 | 6.813 | 21.506 | 43.992 | 7.734 | 66.475 | 35.382 | 21.498 | 42.783 | 28.644 | 31.551 | 37.927 | 20.664 | 17.087 | 33.396 | 63.585 | 20.963 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.092 | 0.257 | -0.294 | -1.831 | -0.019 | 0.516 | -1.103 | 0.085 | -0.447 | 0.651 | 0.129 | -0.074 | -1.047 | 0.468 | 0.384 | -0.533 |
| 02 | -0.354 | -0.112 | -0.054 | -0.643 | 0.564 | 1.133 | -0.885 | 0.189 | -0.079 | -0.038 | 0.159 | -0.471 | -0.543 | -0.336 | -0.343 | -0.276 |
| 03 | -0.133 | 0.046 | 0.419 | -0.652 | 0.076 | 0.701 | -0.662 | 0.725 | -0.163 | -0.186 | -0.293 | -0.204 | -1.13 | 0.087 | -0.059 | -0.273 |
| 04 | -0.594 | 0.335 | -0.071 | -1.408 | 0.608 | 0.285 | -0.311 | 0.082 | -0.835 | 0.344 | -0.03 | -0.094 | -0.495 | 0.059 | 0.362 | 0.057 |
| 05 | -0.802 | 0.464 | -0.153 | -0.627 | -0.292 | 0.928 | -1.968 | 0.578 | -0.535 | 0.063 | 0.104 | 0.27 | -2.695 | 0.166 | -0.298 | 0.268 |
| 06 | -4.4 | -0.425 | -0.154 | -1.123 | 0.413 | 0.902 | -0.468 | 0.569 | -0.526 | 0.029 | -0.225 | -0.842 | -1.285 | 0.734 | 0.534 | -0.14 |
| 07 | -0.803 | -0.64 | 0.101 | -1.174 | 1.166 | 0.564 | -1.177 | -0.047 | -0.047 | -0.179 | 0.298 | -0.161 | -0.547 | 0.215 | 0.132 | -0.836 |
| 08 | -1.253 | 0.003 | 0.043 | 1.05 | -1.268 | 0.406 | -1.963 | 0.891 | -0.683 | 0.032 | -1.124 | 0.713 | -1.93 | -0.433 | -1.678 | 0.449 |
| 09 | -4.4 | -4.4 | -3.227 | 0.142 | -3.246 | -2.285 | -4.4 | 1.394 | -2.978 | -4.4 | -3.236 | 0.466 | -4.4 | -2.622 | -4.4 | 2.175 |
| 10 | -4.4 | -4.4 | -4.4 | -2.557 | -3.136 | -4.4 | -4.4 | -2.033 | -4.4 | -4.4 | -4.4 | -2.707 | -2.286 | -2.236 | -4.4 | 2.732 |
| 11 | -3.174 | -2.271 | -4.4 | -4.4 | -2.948 | -2.708 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.306 | 2.694 | -2.943 | -0.372 |
| 12 | -4.4 | -2.187 | -4.4 | -2.667 | -2.23 | 2.398 | -4.4 | 1.349 | -4.4 | -2.943 | -4.4 | -4.4 | -4.4 | -0.461 | -4.4 | -2.648 |
| 13 | -4.4 | -3.087 | -4.4 | -2.602 | 0.423 | 1.65 | -1.342 | 1.57 | -4.4 | -4.4 | -4.4 | -4.4 | -1.961 | 0.704 | -1.037 | 0.397 |
| 14 | -1.28 | -2.983 | 0.185 | -1.832 | 1.223 | -0.026 | 0.606 | 0.077 | -2.031 | -3.096 | -2.506 | -0.962 | 0.04 | -0.984 | 1.357 | 0.081 |
| 15 | -3.197 | -1.124 | 1.397 | -0.82 | -1.476 | -1.376 | -0.87 | -0.608 | -0.968 | -1.754 | 1.519 | 0.633 | -4.4 | -1.982 | 0.68 | -0.542 |
| 16 | -0.755 | -1.984 | -3.178 | -4.4 | -0.975 | -0.72 | -4.4 | -4.4 | 2.095 | 0.828 | -2.57 | -0.671 | 0.822 | -0.218 | -1.953 | -1.478 |
| 17 | 2.339 | -2.257 | -0.933 | -0.96 | 1.144 | -4.4 | -2.665 | -0.706 | -1.446 | -4.4 | -4.4 | -4.4 | -0.667 | -3.177 | -1.968 | -2.673 |
| 18 | 1.727 | 0.648 | 1.588 | 0.604 | -3.158 | -4.4 | -4.4 | -2.265 | -1.125 | -1.447 | -3.15 | -4.4 | -2.58 | -2.025 | -1.207 | -0.796 |
| 19 | 0.255 | 0.524 | 0.546 | 0.306 | -1.058 | -0.318 | -1.798 | 0.05 | 0.027 | 0.607 | -0.007 | 0.33 | -0.483 | -0.205 | -0.715 | -0.744 |
| 20 | -0.404 | 0.005 | -0.128 | -0.314 | 0.06 | 0.822 | -1.274 | 0.372 | -0.94 | -0.043 | 0.008 | 0.017 | -0.78 | 0.061 | 0.492 | 0.112 |
| 21 | -1.48 | -0.368 | 0.338 | -1.36 | 0.748 | 0.123 | -0.369 | 0.311 | -0.086 | 0.009 | 0.191 | -0.407 | -0.594 | 0.066 | 0.704 | -0.393 |