Model info
| Transcription factor | Cdx4 | ||||||||
| Model | CDX4_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 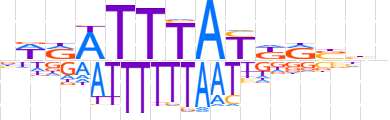 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Consensus | nWRATTTAYKRbhn | ||||||||
| wAUC | 0.8556649397174473 | ||||||||
| Best AUC | 0.8716569321731612 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 513 | ||||||||
| TF family | HOX-related factors{3.1.1} | ||||||||
| TF subfamily | CDX (Caudal type homeobox){3.1.1.9} | ||||||||
| MGI | 88362 | ||||||||
| EntrezGene | 12592 | ||||||||
| UniProt ID | CDX4_MOUSE | ||||||||
| UniProt AC | Q07424 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 49.713 | 2.107 | 17.637 | 72.553 | 57.331 | 0.894 | 1.834 | 58.488 | 35.926 | 5.022 | 9.949 | 89.656 | 15.776 | 2.828 | 18.396 | 61.89 |
| 02 | 27.146 | 9.794 | 93.426 | 28.379 | 6.831 | 0.974 | 3.045 | 0.0 | 4.316 | 4.956 | 35.768 | 2.776 | 48.963 | 3.193 | 180.701 | 49.73 |
| 03 | 70.641 | 2.445 | 0.0 | 14.171 | 10.894 | 0.948 | 0.0 | 7.076 | 251.017 | 7.632 | 1.873 | 52.419 | 64.074 | 4.149 | 0.0 | 12.662 |
| 04 | 6.195 | 0.0 | 2.95 | 387.481 | 0.0 | 0.938 | 0.0 | 14.235 | 0.0 | 0.902 | 0.0 | 0.971 | 0.0 | 0.0 | 0.0 | 86.328 |
| 05 | 0.966 | 0.0 | 0.0 | 5.229 | 0.0 | 0.0 | 0.0 | 1.84 | 0.951 | 0.0 | 0.0 | 1.999 | 7.589 | 1.053 | 6.71 | 473.663 |
| 06 | 0.0 | 0.933 | 0.0 | 8.573 | 0.0 | 0.0 | 0.0 | 1.053 | 0.894 | 0.0 | 2.83 | 2.986 | 13.348 | 21.106 | 15.07 | 433.206 |
| 07 | 14.242 | 0.0 | 0.0 | 0.0 | 22.039 | 0.0 | 0.0 | 0.0 | 17.9 | 0.0 | 0.0 | 0.0 | 445.818 | 0.0 | 0.0 | 0.0 |
| 08 | 18.953 | 104.125 | 3.953 | 372.969 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 1.033 | 0.0 | 11.867 | 6.053 | 36.644 | 1.867 | 26.627 | 38.986 | 1.054 | 0.0 | 0.941 | 1.958 | 43.426 | 4.686 | 209.241 | 115.616 |
| 10 | 14.045 | 2.974 | 58.802 | 6.335 | 3.792 | 0.908 | 0.926 | 0.928 | 41.786 | 16.044 | 152.604 | 38.243 | 29.185 | 3.997 | 110.216 | 19.215 |
| 11 | 2.009 | 44.18 | 25.02 | 17.599 | 2.996 | 13.02 | 1.075 | 6.832 | 25.428 | 197.281 | 55.843 | 43.998 | 2.223 | 24.887 | 25.041 | 12.569 |
| 12 | 9.294 | 7.925 | 9.452 | 5.985 | 63.782 | 105.217 | 4.007 | 106.361 | 27.541 | 51.819 | 14.881 | 12.74 | 9.72 | 27.003 | 22.214 | 22.061 |
| 13 | 25.014 | 25.849 | 42.144 | 17.329 | 58.662 | 52.552 | 14.025 | 66.724 | 2.874 | 17.744 | 18.57 | 11.367 | 14.987 | 44.704 | 43.245 | 44.21 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.46 | -2.54 | -0.563 | 0.835 | 0.601 | -3.206 | -2.656 | 0.621 | 0.138 | -1.766 | -1.119 | 1.046 | -0.672 | -2.286 | -0.521 | 0.677 |
| 02 | -0.139 | -1.134 | 1.087 | -0.095 | -1.478 | -3.145 | -2.221 | -4.4 | -1.906 | -1.778 | 0.133 | -2.303 | 0.445 | -2.179 | 1.745 | 0.46 |
| 03 | 0.809 | -2.413 | -4.4 | -0.776 | -1.031 | -3.165 | -4.4 | -1.444 | 2.073 | -1.372 | -2.638 | 0.512 | 0.712 | -1.942 | -4.4 | -0.886 |
| 04 | -1.57 | -4.4 | -2.249 | 2.506 | -4.4 | -3.172 | -4.4 | -0.772 | -4.4 | -3.2 | -4.4 | -3.147 | -4.4 | -4.4 | -4.4 | 1.008 |
| 05 | -3.151 | -4.4 | -4.4 | -1.728 | -4.4 | -4.4 | -4.4 | -2.653 | -3.162 | -4.4 | -4.4 | -2.584 | -1.378 | -3.089 | -1.494 | 2.707 |
| 06 | -4.4 | -3.176 | -4.4 | -1.261 | -4.4 | -4.4 | -4.4 | -3.089 | -3.206 | -4.4 | -2.286 | -2.238 | -0.834 | -0.387 | -0.716 | 2.618 |
| 07 | -0.771 | -4.4 | -4.4 | -4.4 | -0.344 | -4.4 | -4.4 | -4.4 | -0.548 | -4.4 | -4.4 | -4.4 | 2.646 | -4.4 | -4.4 | -4.4 |
| 08 | -0.492 | 1.195 | -1.986 | 2.468 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -3.103 | -4.4 | -0.948 | -1.592 | 0.157 | -2.641 | -0.158 | 0.219 | -3.088 | -4.4 | -3.169 | -2.601 | 0.326 | -1.83 | 1.891 | 1.299 |
| 10 | -0.785 | -2.242 | 0.626 | -1.549 | -2.024 | -3.195 | -3.181 | -3.18 | 0.287 | -0.655 | 1.576 | 0.2 | -0.068 | -1.976 | 1.252 | -0.479 |
| 11 | -2.58 | 0.343 | -0.219 | -0.565 | -2.235 | -0.859 | -3.074 | -1.477 | -0.203 | 1.832 | 0.575 | 0.339 | -2.495 | -0.225 | -0.218 | -0.893 |
| 12 | -1.184 | -1.337 | -1.168 | -1.602 | 0.707 | 1.205 | -1.974 | 1.216 | -0.125 | 0.501 | -0.729 | -0.88 | -1.141 | -0.144 | -0.336 | -0.343 |
| 13 | -0.22 | -0.187 | 0.296 | -0.58 | 0.624 | 0.515 | -0.786 | 0.752 | -2.272 | -0.557 | -0.512 | -0.99 | -0.722 | 0.354 | 0.321 | 0.343 |