Model info
| Transcription factor | Bcl6 | ||||||||
| Model | BCL6_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 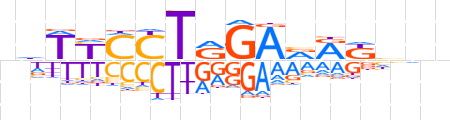 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 16 | ||||||||
| Quality | C | ||||||||
| Consensus | nndMYbTCYAGKRAhn | ||||||||
| wAUC | 0.7353697537191268 | ||||||||
| Best AUC | 0.7759557538066195 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 504 | ||||||||
| TF family | More than 3 adjacent zinc finger factors{2.3.3} | ||||||||
| TF subfamily | BCL6 factors{2.3.3.22} | ||||||||
| MGI | 107187 | ||||||||
| EntrezGene | 12053 | ||||||||
| UniProt ID | BCL6_MOUSE | ||||||||
| UniProt AC | P41183 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 15.031 | 29.05 | 27.047 | 47.864 | 34.605 | 19.674 | 3.957 | 74.188 | 31.957 | 31.692 | 28.756 | 14.045 | 10.175 | 36.963 | 47.209 | 47.789 |
| 02 | 33.378 | 6.786 | 38.556 | 13.047 | 26.989 | 31.464 | 9.656 | 49.27 | 25.655 | 18.701 | 49.787 | 12.825 | 26.479 | 18.281 | 111.851 | 27.274 |
| 03 | 21.532 | 68.085 | 14.92 | 7.964 | 27.174 | 39.889 | 0.0 | 8.169 | 46.286 | 136.62 | 3.792 | 23.152 | 9.031 | 77.486 | 6.701 | 9.198 |
| 04 | 12.258 | 25.555 | 16.177 | 50.034 | 21.467 | 40.637 | 2.94 | 257.038 | 0.938 | 5.779 | 0.0 | 18.696 | 1.983 | 8.319 | 4.187 | 33.994 |
| 05 | 3.05 | 10.441 | 9.651 | 13.503 | 10.068 | 15.162 | 1.015 | 54.046 | 0.945 | 13.411 | 0.933 | 8.015 | 22.746 | 69.158 | 57.208 | 210.649 |
| 06 | 4.78 | 4.957 | 1.002 | 26.07 | 3.898 | 8.698 | 0.945 | 94.631 | 3.946 | 6.019 | 4.042 | 54.8 | 1.897 | 20.223 | 7.125 | 256.968 |
| 07 | 1.101 | 13.42 | 0.0 | 0.0 | 1.937 | 31.944 | 0.0 | 6.016 | 0.0 | 11.998 | 0.0 | 1.115 | 0.0 | 372.088 | 7.999 | 52.382 |
| 08 | 0.0 | 3.038 | 0.0 | 0.0 | 18.901 | 226.004 | 10.718 | 173.827 | 0.0 | 4.948 | 1.029 | 2.023 | 4.196 | 38.665 | 2.971 | 13.682 |
| 09 | 21.091 | 0.0 | 2.006 | 0.0 | 266.566 | 5.017 | 0.0 | 1.071 | 13.777 | 0.94 | 0.0 | 0.0 | 175.247 | 1.833 | 11.426 | 1.026 |
| 10 | 57.271 | 6.474 | 382.61 | 30.326 | 1.897 | 0.0 | 2.946 | 2.947 | 2.219 | 0.0 | 8.784 | 2.43 | 0.0 | 0.0 | 2.096 | 0.0 |
| 11 | 4.929 | 3.041 | 44.817 | 8.601 | 3.301 | 0.0 | 3.173 | 0.0 | 23.698 | 20.108 | 323.282 | 29.348 | 1.014 | 1.097 | 29.579 | 4.013 |
| 12 | 29.168 | 0.0 | 0.972 | 2.802 | 19.006 | 0.0 | 0.0 | 5.239 | 290.768 | 34.226 | 41.424 | 34.434 | 21.359 | 1.948 | 10.378 | 8.276 |
| 13 | 304.996 | 20.292 | 8.579 | 26.433 | 24.425 | 2.028 | 1.017 | 8.705 | 43.91 | 4.823 | 0.0 | 4.04 | 25.999 | 1.885 | 3.291 | 19.577 |
| 14 | 168.115 | 73.738 | 51.879 | 105.598 | 11.647 | 2.924 | 1.007 | 13.449 | 2.885 | 2.822 | 0.914 | 6.266 | 6.801 | 18.773 | 5.034 | 28.147 |
| 15 | 69.899 | 37.076 | 49.432 | 33.042 | 37.319 | 14.281 | 5.129 | 41.528 | 15.21 | 14.047 | 12.871 | 16.705 | 29.539 | 35.207 | 37.294 | 51.42 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.719 | -0.072 | -0.143 | 0.422 | 0.101 | -0.456 | -1.985 | 0.857 | 0.022 | 0.014 | -0.082 | -0.785 | -1.097 | 0.166 | 0.408 | 0.421 |
| 02 | 0.065 | -1.484 | 0.208 | -0.856 | -0.145 | 0.007 | -1.147 | 0.451 | -0.195 | -0.505 | 0.461 | -0.873 | -0.163 | -0.527 | 1.266 | -0.134 |
| 03 | -0.367 | 0.772 | -0.726 | -1.332 | -0.138 | 0.241 | -4.4 | -1.308 | 0.389 | 1.466 | -2.024 | -0.296 | -1.212 | 0.901 | -1.496 | -1.194 |
| 04 | -0.917 | -0.198 | -0.647 | 0.466 | -0.37 | 0.26 | -2.252 | 2.096 | -3.172 | -1.635 | -4.4 | -0.506 | -2.591 | -1.29 | -1.934 | 0.083 |
| 05 | -2.219 | -1.072 | -1.148 | -0.823 | -1.107 | -0.71 | -3.116 | 0.543 | -3.167 | -0.83 | -3.176 | -1.326 | -0.313 | 0.788 | 0.599 | 1.898 |
| 06 | -1.812 | -1.778 | -3.125 | -0.179 | -1.999 | -1.248 | -3.167 | 1.1 | -1.988 | -1.597 | -1.966 | 0.556 | -2.628 | -0.429 | -1.438 | 2.096 |
| 07 | -3.056 | -0.829 | -4.4 | -4.4 | -2.61 | 0.022 | -4.4 | -1.597 | -4.4 | -0.938 | -4.4 | -3.047 | -4.4 | 2.466 | -1.328 | 0.512 |
| 08 | -4.4 | -2.223 | -4.4 | -4.4 | -0.495 | 1.968 | -1.047 | 1.706 | -4.4 | -1.78 | -3.106 | -2.574 | -1.932 | 0.211 | -2.243 | -0.81 |
| 09 | -0.387 | -4.4 | -2.581 | -4.4 | 2.133 | -1.767 | -4.4 | -3.076 | -0.804 | -3.17 | -4.4 | -4.4 | 1.714 | -2.656 | -0.985 | -3.108 |
| 10 | 0.6 | -1.528 | 2.494 | -0.03 | -2.628 | -4.4 | -2.25 | -2.25 | -2.496 | -4.4 | -1.238 | -2.418 | -4.4 | -4.4 | -2.544 | -4.4 |
| 11 | -1.783 | -2.222 | 0.357 | -1.258 | -2.149 | -4.4 | -2.184 | -4.4 | -0.273 | -0.434 | 2.325 | -0.062 | -3.117 | -3.059 | -0.054 | -1.973 |
| 12 | -0.068 | -4.4 | -3.147 | -2.294 | -0.489 | -4.4 | -4.4 | -1.727 | 2.219 | 0.09 | 0.279 | 0.096 | -0.375 | -2.606 | -1.078 | -1.295 |
| 13 | 2.267 | -0.425 | -1.261 | -0.165 | -0.243 | -2.572 | -3.114 | -1.247 | 0.337 | -1.804 | -4.4 | -1.966 | -0.181 | -2.633 | -2.152 | -0.46 |
| 14 | 1.673 | 0.851 | 0.502 | 1.209 | -0.966 | -2.257 | -3.121 | -0.827 | -2.269 | -2.288 | -3.19 | -1.559 | -1.482 | -0.501 | -1.764 | -0.103 |
| 15 | 0.798 | 0.169 | 0.454 | 0.055 | 0.175 | -0.769 | -1.746 | 0.281 | -0.707 | -0.785 | -0.87 | -0.616 | -0.056 | 0.118 | 0.175 | 0.493 |