Model info
| Transcription factor | Neurod1 | ||||||||
| Model | NDF1_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 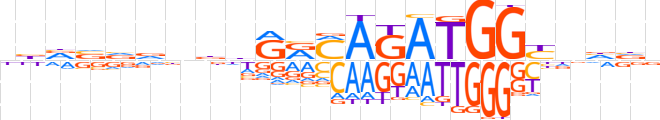 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Consensus | bbvvvvnddRRMAGATGGYnhvv | ||||||||
| wAUC | 0.9092513609210184 | ||||||||
| Best AUC | 0.9092513609210184 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 731 | ||||||||
| TF family | Tal-related factors{1.2.3} | ||||||||
| TF subfamily | Neurogenin / Atonal-like factors{1.2.3.4} | ||||||||
| MGI | 1339708 | ||||||||
| EntrezGene | 18012 | ||||||||
| UniProt ID | NDF1_MOUSE | ||||||||
| UniProt AC | Q60867 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 11.733 | 7.558 | 21.538 | 7.77 | 14.189 | 13.985 | 2.874 | 26.339 | 6.581 | 15.265 | 21.176 | 14.366 | 10.682 | 11.224 | 9.47 | 96.83 |
| 02 | 12.441 | 10.001 | 17.879 | 2.863 | 26.01 | 12.238 | 0.0 | 9.785 | 21.121 | 12.325 | 14.3 | 7.313 | 104.187 | 7.047 | 21.255 | 12.816 |
| 03 | 10.635 | 30.232 | 118.232 | 4.661 | 14.375 | 20.235 | 0.969 | 6.032 | 8.477 | 19.964 | 17.905 | 7.087 | 0.911 | 10.205 | 15.977 | 5.685 |
| 04 | 8.815 | 4.893 | 17.923 | 2.767 | 35.677 | 22.006 | 5.624 | 17.328 | 17.76 | 23.75 | 106.442 | 5.131 | 3.805 | 6.005 | 12.726 | 0.929 |
| 05 | 17.803 | 9.613 | 33.912 | 4.73 | 25.075 | 7.475 | 1.862 | 22.242 | 70.737 | 11.427 | 59.576 | 0.974 | 7.004 | 7.427 | 9.8 | 1.924 |
| 06 | 25.662 | 47.069 | 42.127 | 5.761 | 18.072 | 5.837 | 3.706 | 8.328 | 22.835 | 18.148 | 21.005 | 43.161 | 1.99 | 9.731 | 17.253 | 0.896 |
| 07 | 14.631 | 7.523 | 38.5 | 7.905 | 21.62 | 10.206 | 34.502 | 14.457 | 14.04 | 12.664 | 43.857 | 13.53 | 1.876 | 2.913 | 13.33 | 40.028 |
| 08 | 6.717 | 6.444 | 27.593 | 11.412 | 7.569 | 2.782 | 0.0 | 22.954 | 21.28 | 20.407 | 22.7 | 65.802 | 46.148 | 2.894 | 23.133 | 3.746 |
| 09 | 14.598 | 1.129 | 65.988 | 0.0 | 24.914 | 0.0 | 7.612 | 0.0 | 43.023 | 2.786 | 24.883 | 2.733 | 7.538 | 0.0 | 96.376 | 0.0 |
| 10 | 38.087 | 13.869 | 38.117 | 0.0 | 3.915 | 0.0 | 0.0 | 0.0 | 105.781 | 17.986 | 71.092 | 0.0 | 0.0 | 1.807 | 0.926 | 0.0 |
| 11 | 39.011 | 108.773 | 0.0 | 0.0 | 0.0 | 33.662 | 0.0 | 0.0 | 4.254 | 68.701 | 37.18 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 43.266 | 0.0 | 0.0 | 0.0 | 211.135 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 37.18 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 0.0 | 0.0 | 206.391 | 48.01 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 37.18 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 179.463 | 22.865 | 4.063 | 0.0 | 84.131 | 1.058 | 0.0 | 0.0 |
| 15 | 0.0 | 0.0 | 39.379 | 224.216 | 0.0 | 0.0 | 0.0 | 23.923 | 0.0 | 0.0 | 0.0 | 4.063 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 39.379 | 0.0 | 0.0 | 0.0 | 252.202 | 0.0 |
| 17 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.888 | 0.0 | 282.218 | 8.475 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 0.888 | 0.0 | 0.0 | 0.0 | 0.0 | 15.178 | 140.322 | 15.522 | 111.197 | 0.0 | 1.972 | 0.0 | 6.503 |
| 19 | 1.967 | 4.705 | 7.58 | 0.926 | 46.047 | 24.457 | 44.163 | 27.627 | 4.728 | 3.901 | 3.907 | 2.986 | 11.395 | 16.26 | 44.58 | 46.353 |
| 20 | 25.333 | 12.216 | 19.5 | 7.088 | 18.415 | 11.458 | 3.793 | 15.657 | 59.99 | 19.632 | 7.63 | 12.977 | 45.121 | 7.911 | 13.583 | 11.276 |
| 21 | 18.581 | 4.91 | 118.673 | 6.696 | 21.075 | 10.975 | 5.647 | 13.52 | 5.64 | 15.523 | 17.608 | 5.736 | 6.625 | 13.079 | 11.482 | 15.812 |
| 22 | 11.381 | 8.695 | 20.282 | 11.562 | 12.65 | 17.233 | 1.979 | 12.626 | 23.39 | 14.539 | 102.123 | 13.358 | 8.142 | 7.563 | 22.142 | 3.917 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.43 | -0.853 | 0.164 | -0.827 | -0.245 | -0.259 | -1.75 | 0.362 | -0.985 | -0.173 | 0.147 | -0.233 | -0.521 | -0.473 | -0.637 | 1.655 |
| 02 | -0.373 | -0.584 | -0.019 | -1.753 | 0.35 | -0.389 | -3.958 | -0.606 | 0.145 | -0.382 | -0.237 | -0.885 | 1.728 | -0.92 | 0.151 | -0.344 |
| 03 | -0.525 | 0.499 | 1.854 | -1.31 | -0.232 | 0.103 | -2.642 | -1.068 | -0.744 | 0.09 | -0.017 | -0.915 | -2.686 | -0.565 | -0.129 | -1.124 |
| 04 | -0.706 | -1.264 | -0.016 | -1.784 | 0.662 | 0.185 | -1.134 | -0.049 | -0.025 | 0.26 | 1.749 | -1.22 | -1.497 | -1.072 | -0.351 | -2.672 |
| 05 | -0.023 | -0.623 | 0.612 | -1.296 | 0.314 | -0.864 | -2.126 | 0.196 | 1.342 | -0.455 | 1.171 | -2.637 | -0.926 | -0.87 | -0.604 | -2.099 |
| 06 | 0.337 | 0.937 | 0.827 | -1.111 | -0.008 | -1.099 | -1.521 | -0.761 | 0.222 | -0.004 | 0.139 | 0.851 | -2.07 | -0.611 | -0.054 | -2.698 |
| 07 | -0.215 | -0.858 | 0.738 | -0.811 | 0.168 | -0.565 | 0.629 | -0.227 | -0.255 | -0.356 | 0.867 | -0.291 | -2.12 | -1.738 | -0.306 | 0.776 |
| 08 | -0.966 | -1.005 | 0.408 | -0.457 | -0.852 | -1.779 | -3.958 | 0.227 | 0.152 | 0.111 | 0.216 | 1.27 | 0.917 | -1.744 | 0.234 | -1.511 |
| 09 | -0.217 | -2.528 | 1.273 | -3.958 | 0.308 | -3.958 | -0.847 | -3.958 | 0.848 | -1.777 | 0.306 | -1.794 | -0.856 | -3.958 | 1.65 | -3.958 |
| 10 | 0.727 | -0.267 | 0.728 | -3.958 | -1.47 | -3.958 | -3.958 | -3.958 | 1.743 | -0.013 | 1.347 | -3.958 | -3.958 | -2.151 | -2.675 | -3.958 |
| 11 | 0.751 | 1.771 | -3.958 | -3.958 | -3.958 | 0.605 | -3.958 | -3.958 | -1.394 | 1.313 | 0.703 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 |
| 12 | 0.854 | -3.958 | -3.958 | -3.958 | 2.432 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | 0.703 | -3.958 | -3.958 | -3.958 | -3.958 |
| 13 | -3.958 | -3.958 | 2.409 | 0.957 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | 0.703 |
| 14 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | 2.27 | 0.223 | -1.436 | -3.958 | 1.515 | -2.576 | -3.958 | -3.958 |
| 15 | -3.958 | -3.958 | 0.76 | 2.492 | -3.958 | -3.958 | -3.958 | 0.268 | -3.958 | -3.958 | -3.958 | -1.436 | -3.958 | -3.958 | -3.958 | -3.958 |
| 16 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | 0.76 | -3.958 | -3.958 | -3.958 | 2.61 | -3.958 |
| 17 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -3.958 | -2.705 | -3.958 | 2.722 | -0.744 | -3.958 | -3.958 | -3.958 | -3.958 |
| 18 | -3.958 | -3.958 | -3.958 | -2.705 | -3.958 | -3.958 | -3.958 | -3.958 | -0.179 | 2.024 | -0.157 | 1.792 | -3.958 | -2.078 | -3.958 | -0.997 |
| 19 | -2.08 | -1.301 | -0.851 | -2.675 | 0.915 | 0.289 | 0.874 | 0.41 | -1.296 | -1.474 | -1.472 | -1.716 | -0.458 | -0.112 | 0.883 | 0.922 |
| 20 | 0.324 | -0.391 | 0.066 | -0.915 | 0.01 | -0.453 | -1.499 | -0.149 | 1.178 | 0.073 | -0.844 | -0.332 | 0.895 | -0.81 | -0.287 | -0.468 |
| 21 | 0.019 | -1.261 | 1.857 | -0.969 | 0.143 | -0.495 | -1.13 | -0.292 | -1.131 | -0.157 | -0.034 | -1.115 | -0.979 | -0.324 | -0.451 | -0.139 |
| 22 | -0.459 | -0.719 | 0.105 | -0.444 | -0.357 | -0.055 | -2.074 | -0.359 | 0.245 | -0.221 | 1.708 | -0.304 | -0.782 | -0.853 | 0.191 | -1.47 |