Model info
| Transcription factor | Stat1 | ||||||||
| Model | STAT1_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 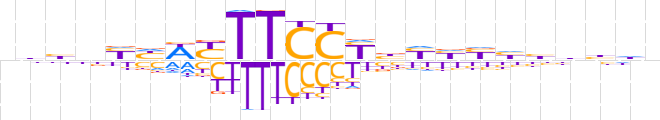 | ||||||||
| LOGO (reverse complement) | 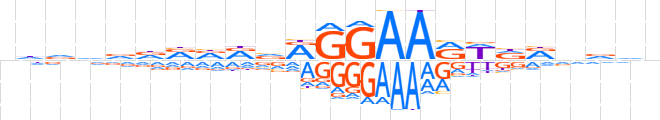 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nbhnYhWYTTCCYbYhbhhhhdn | ||||||||
| wAUC | 0.7906657931512691 | ||||||||
| Best AUC | 0.818755033918522 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT1{6.2.1.0.1} | ||||||||
| MGI | 103063 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | STAT1_MOUSE | ||||||||
| UniProt AC | P42225 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 11.06 | 33.908 | 23.816 | 36.999 | 19.856 | 36.801 | 5.743 | 63.117 | 5.27 | 23.296 | 33.909 | 42.505 | 14.676 | 36.617 | 67.029 | 45.397 |
| 02 | 8.752 | 11.744 | 10.706 | 19.66 | 17.332 | 43.195 | 1.133 | 68.962 | 25.045 | 21.786 | 17.273 | 66.394 | 16.887 | 46.489 | 36.172 | 88.47 |
| 03 | 12.608 | 16.118 | 26.948 | 12.341 | 36.004 | 40.413 | 4.999 | 41.797 | 14.482 | 16.917 | 25.772 | 8.113 | 18.74 | 52.783 | 50.784 | 121.18 |
| 04 | 9.139 | 23.152 | 17.528 | 32.015 | 16.292 | 24.039 | 1.23 | 84.67 | 4.787 | 26.098 | 8.197 | 69.421 | 20.027 | 28.424 | 17.879 | 117.102 |
| 05 | 18.291 | 15.852 | 6.066 | 10.036 | 26.321 | 55.729 | 3.818 | 15.845 | 10.117 | 25.096 | 2.141 | 7.481 | 26.497 | 184.841 | 42.123 | 49.747 |
| 06 | 64.751 | 2.027 | 2.614 | 11.834 | 200.909 | 22.854 | 10.033 | 47.721 | 32.366 | 7.198 | 9.609 | 4.975 | 35.448 | 2.091 | 16.31 | 29.26 |
| 07 | 6.425 | 177.044 | 29.731 | 120.275 | 1.195 | 22.121 | 2.164 | 8.691 | 0.0 | 15.073 | 2.167 | 21.326 | 0.0 | 56.987 | 7.475 | 29.327 |
| 08 | 0.0 | 0.0 | 0.0 | 7.62 | 12.249 | 5.401 | 0.0 | 253.576 | 1.962 | 1.123 | 1.057 | 37.395 | 1.899 | 3.194 | 0.0 | 174.526 |
| 09 | 0.0 | 0.0 | 2.489 | 13.621 | 1.24 | 2.921 | 0.0 | 5.556 | 0.0 | 0.0 | 1.057 | 0.0 | 3.122 | 4.064 | 6.763 | 459.168 |
| 10 | 0.0 | 4.361 | 0.0 | 0.0 | 0.0 | 5.137 | 0.0 | 1.848 | 0.0 | 8.986 | 0.0 | 1.323 | 4.226 | 394.708 | 1.021 | 78.391 |
| 11 | 0.0 | 0.0 | 0.0 | 4.226 | 0.0 | 334.949 | 0.0 | 78.243 | 0.0 | 0.0 | 0.0 | 1.021 | 0.778 | 51.451 | 0.0 | 29.333 |
| 12 | 0.0 | 0.0 | 0.0 | 0.778 | 33.963 | 70.798 | 4.262 | 277.378 | 0.0 | 0.0 | 0.0 | 0.0 | 5.504 | 16.071 | 25.474 | 65.774 |
| 13 | 3.151 | 14.097 | 15.157 | 7.061 | 7.481 | 47.221 | 4.229 | 27.937 | 1.291 | 18.864 | 2.644 | 6.937 | 23.844 | 126.01 | 72.647 | 121.428 |
| 14 | 5.434 | 12.092 | 0.712 | 17.53 | 47.451 | 38.45 | 0.0 | 120.291 | 8.868 | 21.81 | 3.969 | 60.029 | 18.282 | 48.067 | 9.435 | 87.58 |
| 15 | 10.73 | 11.094 | 15.731 | 42.479 | 14.466 | 36.886 | 2.033 | 67.035 | 3.989 | 3.202 | 0.0 | 6.923 | 30.545 | 50.384 | 34.864 | 169.638 |
| 16 | 10.946 | 7.212 | 10.021 | 31.551 | 12.169 | 27.177 | 1.104 | 61.116 | 10.118 | 16.933 | 9.53 | 16.046 | 8.353 | 91.686 | 22.648 | 163.388 |
| 17 | 16.029 | 8.002 | 3.052 | 14.504 | 17.832 | 61.656 | 1.174 | 62.346 | 8.34 | 12.867 | 6.584 | 15.512 | 11.467 | 98.78 | 30.186 | 131.669 |
| 18 | 11.334 | 19.047 | 7.643 | 15.643 | 28.62 | 76.069 | 2.673 | 73.941 | 14.178 | 4.275 | 9.496 | 13.048 | 23.571 | 79.734 | 24.718 | 96.008 |
| 19 | 27.775 | 17.952 | 17.061 | 14.915 | 30.709 | 74.721 | 0.975 | 72.721 | 14.345 | 10.25 | 7.798 | 12.137 | 16.199 | 43.756 | 47.752 | 90.935 |
| 20 | 13.116 | 37.057 | 18.314 | 20.541 | 41.267 | 66.088 | 0.0 | 39.324 | 19.215 | 17.259 | 11.579 | 25.532 | 20.771 | 74.19 | 20.21 | 75.536 |
| 21 | 24.815 | 18.828 | 35.81 | 14.917 | 85.535 | 19.544 | 7.712 | 81.803 | 26.113 | 7.603 | 0.0 | 16.387 | 33.568 | 15.965 | 38.306 | 73.094 |
| 22 | 58.609 | 28.334 | 50.36 | 32.727 | 26.734 | 25.095 | 1.061 | 9.049 | 30.255 | 22.583 | 17.129 | 11.861 | 15.241 | 61.309 | 50.297 | 59.355 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.016 | 0.081 | -0.268 | 0.167 | -0.447 | 0.162 | -1.641 | 0.697 | -1.721 | -0.29 | 0.081 | 0.304 | -0.742 | 0.157 | 0.757 | 0.37 |
| 02 | -1.242 | -0.958 | -1.048 | -0.456 | -0.58 | 0.32 | -3.035 | 0.785 | -0.218 | -0.355 | -0.583 | 0.747 | -0.605 | 0.393 | 0.145 | 1.033 |
| 03 | -0.89 | -0.651 | -0.146 | -0.91 | 0.14 | 0.254 | -1.77 | 0.288 | -0.755 | -0.603 | -0.19 | -1.314 | -0.503 | 0.519 | 0.481 | 1.346 |
| 04 | -1.2 | -0.296 | -0.569 | 0.024 | -0.64 | -0.259 | -2.973 | 0.989 | -1.811 | -0.178 | -1.304 | 0.791 | -0.438 | -0.094 | -0.549 | 1.312 |
| 05 | -0.527 | -0.667 | -1.59 | -1.11 | -0.169 | 0.573 | -2.018 | -0.667 | -1.102 | -0.216 | -2.527 | -1.391 | -0.163 | 1.767 | 0.295 | 0.46 |
| 06 | 0.722 | -2.572 | -2.355 | -0.951 | 1.85 | -0.308 | -1.11 | 0.419 | 0.035 | -1.428 | -1.152 | -1.775 | 0.125 | -2.546 | -0.639 | -0.065 |
| 07 | -1.536 | 1.724 | -0.049 | 1.339 | -2.995 | -0.34 | -2.517 | -1.248 | -4.4 | -0.716 | -2.516 | -0.376 | -4.4 | 0.595 | -1.392 | -0.063 |
| 08 | -4.4 | -4.4 | -4.4 | -1.374 | -0.918 | -1.698 | -4.4 | 2.083 | -2.6 | -3.042 | -3.086 | 0.178 | -2.627 | -2.178 | -4.4 | 1.71 |
| 09 | -4.4 | -4.4 | -2.397 | -0.815 | -2.967 | -2.258 | -4.4 | -1.672 | -4.4 | -4.4 | -3.086 | -4.4 | -2.199 | -1.961 | -1.487 | 2.676 |
| 10 | -4.4 | -1.896 | -4.4 | -4.4 | -4.4 | -1.745 | -4.4 | -2.649 | -4.4 | -1.216 | -4.4 | -2.917 | -1.925 | 2.525 | -3.111 | 0.912 |
| 11 | -4.4 | -4.4 | -4.4 | -1.925 | -4.4 | 2.361 | -4.4 | 0.91 | -4.4 | -4.4 | -4.4 | -3.111 | -3.301 | 0.494 | -4.4 | -0.063 |
| 12 | -4.4 | -4.4 | -4.4 | -3.301 | 0.082 | 0.811 | -1.918 | 2.172 | -4.4 | -4.4 | -4.4 | -4.4 | -1.681 | -0.654 | -0.202 | 0.738 |
| 13 | -2.19 | -0.781 | -0.711 | -1.446 | -1.391 | 0.409 | -1.925 | -0.111 | -2.936 | -0.497 | -2.345 | -1.463 | -0.267 | 1.385 | 0.837 | 1.348 |
| 14 | -1.693 | -0.93 | -3.359 | -0.569 | 0.413 | 0.205 | -4.4 | 1.339 | -1.229 | -0.354 | -1.983 | 0.647 | -0.527 | 0.426 | -1.17 | 1.023 |
| 15 | -1.046 | -1.014 | -0.674 | 0.304 | -0.756 | 0.164 | -2.57 | 0.757 | -1.978 | -2.176 | -4.4 | -1.465 | -0.023 | 0.473 | 0.108 | 1.682 |
| 16 | -1.027 | -1.426 | -1.112 | 0.009 | -0.924 | -0.138 | -3.054 | 0.665 | -1.102 | -0.602 | -1.16 | -0.655 | -1.286 | 1.068 | -0.317 | 1.644 |
| 17 | -0.656 | -1.327 | -2.219 | -0.754 | -0.552 | 0.673 | -3.008 | 0.685 | -1.288 | -0.87 | -1.512 | -0.688 | -0.982 | 1.142 | -0.034 | 1.429 |
| 18 | -0.993 | -0.487 | -1.371 | -0.68 | -0.087 | 0.882 | -2.336 | 0.854 | -0.776 | -1.915 | -1.163 | -0.856 | -0.278 | 0.929 | -0.231 | 1.114 |
| 19 | -0.116 | -0.545 | -0.595 | -0.726 | -0.017 | 0.865 | -3.144 | 0.838 | -0.764 | -1.09 | -1.352 | -0.927 | -0.646 | 0.333 | 0.42 | 1.06 |
| 20 | -0.851 | 0.169 | -0.526 | -0.413 | 0.275 | 0.742 | -4.4 | 0.227 | -0.479 | -0.584 | -0.972 | -0.199 | -0.402 | 0.857 | -0.429 | 0.875 |
| 21 | -0.227 | -0.499 | 0.135 | -0.726 | 0.999 | -0.462 | -1.363 | 0.955 | -0.177 | -1.376 | -4.4 | -0.634 | 0.071 | -0.66 | 0.201 | 0.843 |
| 22 | 0.623 | -0.097 | 0.473 | 0.046 | -0.154 | -0.216 | -3.083 | -1.21 | -0.032 | -0.32 | -0.591 | -0.949 | -0.705 | 0.668 | 0.471 | 0.636 |