Model info
| Transcription factor | Evx1 | ||||||||
| Model | EVX1_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 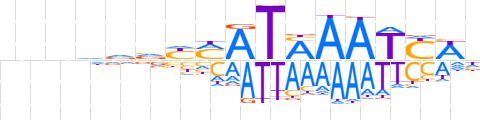 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 17 | ||||||||
| Quality | C | ||||||||
| Consensus | nWRATTKATKRbhhnnn | ||||||||
| wAUC | 0.8342128522087124 | ||||||||
| Best AUC | 0.8342128522087123 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | HOX-related factors{3.1.1} | ||||||||
| TF subfamily | EVX (Even-skipped homolog){3.1.1.10} | ||||||||
| MGI | 95461 | ||||||||
| EntrezGene | 14028 | ||||||||
| UniProt ID | EVX1_MOUSE | ||||||||
| UniProt AC | P23683 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 43.548 | 5.984 | 11.982 | 127.124 | 29.19 | 2.804 | 0.0 | 52.454 | 13.585 | 2.068 | 15.68 | 79.144 | 14.005 | 5.041 | 15.974 | 81.418 |
| 02 | 5.29 | 5.851 | 80.992 | 8.194 | 10.055 | 0.928 | 0.991 | 3.924 | 7.755 | 3.014 | 23.413 | 9.452 | 41.994 | 12.582 | 266.255 | 19.309 |
| 03 | 43.089 | 6.854 | 0.0 | 15.151 | 12.126 | 0.0 | 0.0 | 10.248 | 321.353 | 7.015 | 2.811 | 40.474 | 14.817 | 1.111 | 0.0 | 24.952 |
| 04 | 22.257 | 1.771 | 15.534 | 351.824 | 0.948 | 0.0 | 0.0 | 14.031 | 0.0 | 0.0 | 0.0 | 2.811 | 7.017 | 0.961 | 0.0 | 82.846 |
| 05 | 6.848 | 0.0 | 1.06 | 22.313 | 0.961 | 0.0 | 0.0 | 1.771 | 1.86 | 0.0 | 0.0 | 13.673 | 25.611 | 1.018 | 8.495 | 416.389 |
| 06 | 2.104 | 0.955 | 11.177 | 21.045 | 0.0 | 1.018 | 0.0 | 0.0 | 4.252 | 0.0 | 2.221 | 3.082 | 25.387 | 20.893 | 64.674 | 343.192 |
| 07 | 30.838 | 0.0 | 0.905 | 0.0 | 22.865 | 0.0 | 0.0 | 0.0 | 76.24 | 1.833 | 0.0 | 0.0 | 356.72 | 7.674 | 1.923 | 1.002 |
| 08 | 9.626 | 60.654 | 10.03 | 406.353 | 0.0 | 1.881 | 0.0 | 7.626 | 0.0 | 0.905 | 0.0 | 1.923 | 0.0 | 0.0 | 0.0 | 1.002 |
| 09 | 2.999 | 0.0 | 2.882 | 3.745 | 16.68 | 0.0 | 15.736 | 31.024 | 1.938 | 0.0 | 1.999 | 6.093 | 62.624 | 0.0 | 254.257 | 100.023 |
| 10 | 11.782 | 0.948 | 65.652 | 5.86 | 0.0 | 0.0 | 0.0 | 0.0 | 86.44 | 20.901 | 127.831 | 39.701 | 30.014 | 5.662 | 91.535 | 13.674 |
| 11 | 5.781 | 65.519 | 35.094 | 21.842 | 7.694 | 7.149 | 0.0 | 12.668 | 20.923 | 141.977 | 53.915 | 68.203 | 7.986 | 22.728 | 13.815 | 14.706 |
| 12 | 7.802 | 17.918 | 3.071 | 13.593 | 40.454 | 93.865 | 7.068 | 95.987 | 14.019 | 39.583 | 24.387 | 24.836 | 14.592 | 41.403 | 19.168 | 42.256 |
| 13 | 11.001 | 26.991 | 26.906 | 11.968 | 74.652 | 33.223 | 3.768 | 81.126 | 7.788 | 14.581 | 11.398 | 19.926 | 19.249 | 32.947 | 31.282 | 93.194 |
| 14 | 21.244 | 20.0 | 37.053 | 34.391 | 36.865 | 20.586 | 6.947 | 43.343 | 10.746 | 17.607 | 18.591 | 26.412 | 24.68 | 55.875 | 56.46 | 69.2 |
| 15 | 21.045 | 16.154 | 32.542 | 23.795 | 29.073 | 32.244 | 5.138 | 47.613 | 24.172 | 16.415 | 31.498 | 46.965 | 22.053 | 30.721 | 56.441 | 64.132 |
| 16 | 29.693 | 24.712 | 31.201 | 10.736 | 29.917 | 18.898 | 3.093 | 43.626 | 33.759 | 27.963 | 27.682 | 36.216 | 18.923 | 33.702 | 57.502 | 72.378 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.328 | -1.602 | -0.939 | 1.394 | -0.067 | -2.294 | -4.4 | 0.513 | -0.817 | -2.556 | -0.678 | 0.922 | -0.788 | -1.763 | -0.659 | 0.95 |
| 02 | -1.718 | -1.624 | 0.945 | -1.305 | -1.108 | -3.18 | -3.133 | -1.993 | -1.357 | -2.23 | -0.285 | -1.168 | 0.292 | -0.892 | 2.132 | -0.474 |
| 03 | 0.318 | -1.474 | -4.4 | -0.711 | -0.927 | -4.4 | -4.4 | -1.09 | 2.319 | -1.452 | -2.291 | 0.256 | -0.733 | -3.049 | -4.4 | -0.222 |
| 04 | -0.334 | -2.684 | -0.687 | 2.41 | -3.164 | -4.4 | -4.4 | -0.786 | -4.4 | -4.4 | -4.4 | -2.291 | -1.452 | -3.154 | -4.4 | 0.967 |
| 05 | -1.475 | -4.4 | -3.084 | -0.332 | -3.154 | -4.4 | -4.4 | -2.684 | -2.644 | -4.4 | -4.4 | -0.811 | -0.196 | -3.113 | -1.27 | 2.578 |
| 06 | -2.541 | -3.159 | -1.006 | -0.389 | -4.4 | -3.113 | -4.4 | -4.4 | -1.92 | -4.4 | -2.495 | -2.21 | -0.205 | -0.397 | 0.721 | 2.385 |
| 07 | -0.013 | -4.4 | -3.197 | -4.4 | -0.308 | -4.4 | -4.4 | -4.4 | 0.885 | -2.656 | -4.4 | -4.4 | 2.424 | -1.367 | -2.617 | -3.125 |
| 08 | -1.15 | 0.657 | -1.111 | 2.554 | -4.4 | -2.635 | -4.4 | -1.373 | -4.4 | -3.197 | -4.4 | -2.617 | -4.4 | -4.4 | -4.4 | -3.125 |
| 09 | -2.234 | -4.4 | -2.269 | -2.035 | -0.617 | -4.4 | -0.674 | -0.007 | -2.61 | -4.4 | -2.584 | -1.585 | 0.689 | -4.4 | 2.085 | 1.155 |
| 10 | -0.955 | -3.164 | 0.736 | -1.622 | -4.4 | -4.4 | -4.4 | -4.4 | 1.01 | -0.396 | 1.399 | 0.237 | -0.04 | -1.654 | 1.067 | -0.811 |
| 11 | -1.635 | 0.734 | 0.115 | -0.353 | -1.365 | -1.435 | -4.4 | -0.885 | -0.395 | 1.504 | 0.54 | 0.774 | -1.329 | -0.314 | -0.801 | -0.74 |
| 12 | -1.351 | -0.547 | -2.213 | -0.817 | 0.255 | 1.092 | -1.445 | 1.114 | -0.787 | 0.234 | -0.245 | -0.227 | -0.748 | 0.278 | -0.481 | 0.299 |
| 13 | -1.022 | -0.145 | -0.148 | -0.94 | 0.864 | 0.06 | -2.03 | 0.946 | -1.353 | -0.748 | -0.987 | -0.443 | -0.477 | 0.052 | 0.001 | 1.084 |
| 14 | -0.38 | -0.439 | 0.168 | 0.095 | 0.163 | -0.411 | -1.462 | 0.324 | -1.044 | -0.564 | -0.511 | -0.166 | -0.233 | 0.576 | 0.586 | 0.788 |
| 15 | -0.389 | -0.648 | 0.04 | -0.269 | -0.071 | 0.031 | -1.745 | 0.417 | -0.253 | -0.633 | 0.008 | 0.403 | -0.343 | -0.017 | 0.586 | 0.713 |
| 16 | -0.05 | -0.231 | -0.002 | -1.045 | -0.043 | -0.495 | -2.207 | 0.33 | 0.076 | -0.11 | -0.12 | 0.146 | -0.494 | 0.075 | 0.604 | 0.833 |