Model info
| Transcription factor | Jund | ||||||||
| Model | JUND_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 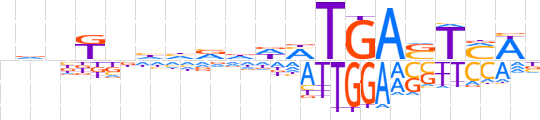 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | C | ||||||||
| Consensus | ndnKbhhvdWMTGASTMWn | ||||||||
| wAUC | 0.8658295450293818 | ||||||||
| Best AUC | 0.8658295450293818 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | Jun-related factors{1.1.1} | ||||||||
| TF subfamily | Jun factors{1.1.1.1} | ||||||||
| MGI | 96648 | ||||||||
| EntrezGene | 16478 | ||||||||
| UniProt ID | JUND_MOUSE | ||||||||
| UniProt AC | P15066 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 74.1 | 15.769 | 47.051 | 20.726 | 67.877 | 7.314 | 5.884 | 32.976 | 35.082 | 7.013 | 19.268 | 27.278 | 31.402 | 21.727 | 57.764 | 28.77 |
| 02 | 32.727 | 49.891 | 60.955 | 64.888 | 12.458 | 14.149 | 0.0 | 25.215 | 20.201 | 30.391 | 36.586 | 42.79 | 9.254 | 25.564 | 31.933 | 42.999 |
| 03 | 0.0 | 3.087 | 42.897 | 28.656 | 2.729 | 2.29 | 7.098 | 107.879 | 4.95 | 2.824 | 37.12 | 84.579 | 0.0 | 1.068 | 100.651 | 74.173 |
| 04 | 3.838 | 0.871 | 2.97 | 0.0 | 4.282 | 3.215 | 0.856 | 0.916 | 21.346 | 11.246 | 133.457 | 21.716 | 41.562 | 83.291 | 28.877 | 141.558 |
| 05 | 10.53 | 42.32 | 8.875 | 9.303 | 19.848 | 34.202 | 1.829 | 42.744 | 131.017 | 16.075 | 7.938 | 11.129 | 11.859 | 39.806 | 7.409 | 105.116 |
| 06 | 123.145 | 21.051 | 16.928 | 12.129 | 57.738 | 23.525 | 0.0 | 51.139 | 17.953 | 3.106 | 0.925 | 4.068 | 26.492 | 89.493 | 19.278 | 33.029 |
| 07 | 115.136 | 19.401 | 82.351 | 8.44 | 90.662 | 14.614 | 10.031 | 21.869 | 14.679 | 0.929 | 19.547 | 1.977 | 10.54 | 9.809 | 77.146 | 2.87 |
| 08 | 84.263 | 38.212 | 60.504 | 48.037 | 21.413 | 5.099 | 0.918 | 17.322 | 118.469 | 13.499 | 20.276 | 36.832 | 13.677 | 1.043 | 14.278 | 6.158 |
| 09 | 120.866 | 3.616 | 50.266 | 63.074 | 18.699 | 2.161 | 1.852 | 35.141 | 47.517 | 2.092 | 15.674 | 30.693 | 9.762 | 0.0 | 15.101 | 83.486 |
| 10 | 162.722 | 12.534 | 16.606 | 4.981 | 7.869 | 0.0 | 0.0 | 0.0 | 61.273 | 7.654 | 8.042 | 5.924 | 89.967 | 47.166 | 22.401 | 52.86 |
| 11 | 1.008 | 0.0 | 0.889 | 319.934 | 2.208 | 0.0 | 0.0 | 65.146 | 0.0 | 0.0 | 0.0 | 47.049 | 0.0 | 0.0 | 0.0 | 63.765 |
| 12 | 0.0 | 0.0 | 3.216 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.889 | 0.0 | 0.0 | 0.0 | 1.897 | 5.008 | 443.091 | 45.899 |
| 13 | 2.786 | 0.0 | 0.0 | 0.0 | 4.098 | 0.0 | 0.0 | 0.91 | 432.887 | 2.787 | 5.589 | 5.044 | 43.986 | 0.965 | 0.948 | 0.0 |
| 14 | 37.567 | 232.196 | 204.059 | 9.935 | 1.896 | 0.0 | 1.856 | 0.0 | 0.981 | 4.687 | 0.868 | 0.0 | 0.0 | 1.044 | 4.91 | 0.0 |
| 15 | 3.266 | 2.072 | 3.108 | 31.998 | 29.688 | 4.894 | 3.892 | 199.454 | 7.923 | 9.853 | 2.662 | 191.256 | 0.0 | 0.0 | 2.037 | 7.897 |
| 16 | 12.15 | 13.539 | 6.798 | 8.389 | 7.975 | 6.975 | 0.0 | 1.869 | 2.0 | 3.867 | 0.0 | 5.832 | 63.795 | 318.319 | 35.087 | 13.404 |
| 17 | 55.413 | 2.032 | 13.155 | 15.321 | 308.963 | 6.134 | 3.843 | 23.76 | 13.957 | 3.699 | 6.799 | 17.429 | 2.062 | 19.388 | 4.017 | 4.028 |
| 18 | 46.461 | 87.469 | 111.919 | 134.546 | 22.233 | 1.923 | 0.0 | 7.098 | 9.257 | 3.909 | 12.652 | 1.997 | 11.001 | 8.796 | 23.465 | 17.276 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.856 | -0.672 | 0.405 | -0.404 | 0.769 | -1.413 | -1.618 | 0.053 | 0.114 | -1.453 | -0.476 | -0.134 | 0.005 | -0.358 | 0.609 | -0.082 |
| 02 | 0.046 | 0.463 | 0.662 | 0.724 | -0.901 | -0.778 | -4.4 | -0.212 | -0.43 | -0.028 | 0.156 | 0.311 | -1.188 | -0.198 | 0.021 | 0.316 |
| 03 | -4.4 | -2.209 | 0.313 | -0.086 | -2.318 | -2.469 | -1.441 | 1.23 | -1.779 | -2.287 | 0.17 | 0.988 | -4.4 | -3.079 | 1.161 | 0.857 |
| 04 | -2.013 | -3.224 | -2.243 | -4.4 | -1.913 | -2.173 | -3.236 | -3.189 | -0.375 | -1.0 | 1.442 | -0.359 | 0.282 | 0.973 | -0.078 | 1.501 |
| 05 | -1.064 | 0.3 | -1.228 | -1.183 | -0.447 | 0.089 | -2.658 | 0.31 | 1.424 | -0.653 | -1.335 | -1.01 | -0.949 | 0.239 | -1.401 | 1.204 |
| 06 | 1.362 | -0.389 | -0.603 | -0.927 | 0.608 | -0.28 | -4.4 | 0.488 | -0.545 | -2.203 | -3.182 | -1.96 | -0.163 | 1.044 | -0.475 | 0.055 |
| 07 | 1.295 | -0.469 | 0.961 | -1.276 | 1.057 | -0.746 | -1.111 | -0.352 | -0.742 | -3.179 | -0.462 | -2.593 | -1.063 | -1.132 | 0.896 | -2.273 |
| 08 | 0.984 | 0.199 | 0.655 | 0.426 | -0.372 | -1.752 | -3.187 | -0.58 | 1.324 | -0.823 | -0.426 | 0.162 | -0.811 | -3.096 | -0.769 | -1.575 |
| 09 | 1.344 | -2.067 | 0.471 | 0.696 | -0.505 | -2.519 | -2.648 | 0.116 | 0.415 | -2.546 | -0.678 | -0.018 | -1.137 | -4.4 | -0.714 | 0.975 |
| 10 | 1.64 | -0.895 | -0.621 | -1.774 | -1.343 | -4.4 | -4.4 | -4.4 | 0.667 | -1.37 | -1.323 | -1.612 | 1.049 | 0.408 | -0.328 | 0.521 |
| 11 | -3.121 | -4.4 | -3.209 | 2.315 | -2.5 | -4.4 | -4.4 | 0.728 | -4.4 | -4.4 | -4.4 | 0.405 | -4.4 | -4.4 | -4.4 | 0.707 |
| 12 | -4.4 | -4.4 | -2.172 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.209 | -4.4 | -4.4 | -4.4 | -2.628 | -1.769 | 2.64 | 0.38 |
| 13 | -2.299 | -4.4 | -4.4 | -4.4 | -1.953 | -4.4 | -4.4 | -3.193 | 2.617 | -2.299 | -1.666 | -1.762 | 0.338 | -3.152 | -3.165 | -4.4 |
| 14 | 0.182 | 1.995 | 1.866 | -1.12 | -2.628 | -4.4 | -2.646 | -4.4 | -3.14 | -1.83 | -3.226 | -4.4 | -4.4 | -3.095 | -1.787 | -4.4 |
| 15 | -2.158 | -2.554 | -2.203 | 0.023 | -0.051 | -1.79 | -2.0 | 1.843 | -1.337 | -1.128 | -2.339 | 1.801 | -4.4 | -4.4 | -2.568 | -1.34 |
| 16 | -0.926 | -0.82 | -1.482 | -1.282 | -1.33 | -1.458 | -4.4 | -2.64 | -2.584 | -2.006 | -4.4 | -1.627 | 0.707 | 2.31 | 0.114 | -0.83 |
| 17 | 0.567 | -2.57 | -0.848 | -0.7 | 2.28 | -1.579 | -2.012 | -0.27 | -0.791 | -2.046 | -1.482 | -0.574 | -2.558 | -0.47 | -1.972 | -1.969 |
| 18 | 0.393 | 1.021 | 1.267 | 1.45 | -0.335 | -2.616 | -4.4 | -1.441 | -1.188 | -1.996 | -0.886 | -2.585 | -1.022 | -1.237 | -0.282 | -0.583 |