Model info
| Transcription factor | Cebpb | ||||||||
| Model | CEBPB_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 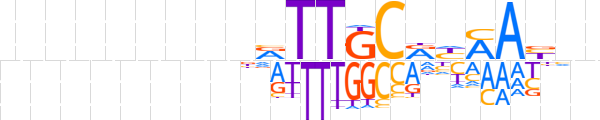 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Consensus | nnRTKRYGCAAYhnnnnnnnn | ||||||||
| wAUC | 0.9303774581142188 | ||||||||
| Best AUC | 0.9474797264071769 | ||||||||
| Benchmark datasets | 10 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | C/EBP-related{1.1.8} | ||||||||
| TF subfamily | C/EBP{1.1.8.1} | ||||||||
| MGI | 88373 | ||||||||
| EntrezGene | 12608 | ||||||||
| UniProt ID | CEBPB_MOUSE | ||||||||
| UniProt AC | P28033 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 32.157 | 17.756 | 53.618 | 26.649 | 19.466 | 22.92 | 3.514 | 50.546 | 32.461 | 25.363 | 46.385 | 47.427 | 5.755 | 18.495 | 47.053 | 50.433 |
| 02 | 37.541 | 8.917 | 41.51 | 1.87 | 41.996 | 26.258 | 15.351 | 0.929 | 90.212 | 27.213 | 29.99 | 3.155 | 65.839 | 35.033 | 74.183 | 0.0 |
| 03 | 0.0 | 1.3 | 0.0 | 234.289 | 0.0 | 0.0 | 0.0 | 97.422 | 0.0 | 0.0 | 2.859 | 158.175 | 0.0 | 0.0 | 0.0 | 5.954 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 0.0 | 1.123 | 1.737 | 2.211 | 1.919 | 180.869 | 310.841 |
| 05 | 1.103 | 1.108 | 0.0 | 0.0 | 0.858 | 1.062 | 0.0 | 0.0 | 133.97 | 1.705 | 7.913 | 38.403 | 49.689 | 1.763 | 216.639 | 45.787 |
| 06 | 16.399 | 31.829 | 8.518 | 128.873 | 0.0 | 0.0 | 0.0 | 5.637 | 38.632 | 62.421 | 7.751 | 115.748 | 3.526 | 29.345 | 1.756 | 49.565 |
| 07 | 0.0 | 0.0 | 58.556 | 0.0 | 0.0 | 0.0 | 121.882 | 1.714 | 0.0 | 0.0 | 18.025 | 0.0 | 0.831 | 0.0 | 298.011 | 0.981 |
| 08 | 0.0 | 0.831 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 63.767 | 402.04 | 0.0 | 30.667 | 0.0 | 2.695 | 0.0 | 0.0 |
| 09 | 61.876 | 1.891 | 0.0 | 0.0 | 405.566 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 30.667 | 0.0 | 0.0 | 0.0 |
| 10 | 495.853 | 0.0 | 2.256 | 0.0 | 1.891 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 19.678 | 148.437 | 55.518 | 274.111 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.256 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 4.122 | 8.601 | 1.954 | 5.002 | 57.869 | 42.348 | 1.955 | 46.266 | 31.335 | 14.652 | 8.57 | 0.961 | 81.777 | 91.327 | 61.742 | 41.522 |
| 13 | 24.408 | 49.249 | 70.152 | 31.294 | 46.685 | 44.686 | 19.885 | 45.671 | 14.962 | 25.7 | 11.947 | 21.611 | 11.441 | 31.836 | 29.084 | 21.389 |
| 14 | 22.626 | 22.257 | 29.385 | 23.229 | 52.145 | 31.234 | 14.193 | 53.899 | 29.571 | 42.257 | 27.407 | 31.834 | 21.504 | 35.264 | 29.741 | 33.455 |
| 15 | 19.587 | 33.011 | 52.968 | 20.279 | 35.353 | 45.422 | 5.543 | 44.694 | 15.789 | 42.14 | 24.04 | 18.757 | 16.434 | 42.494 | 44.799 | 38.69 |
| 16 | 16.705 | 11.826 | 38.592 | 20.04 | 41.739 | 43.506 | 9.829 | 67.993 | 32.034 | 23.813 | 42.661 | 28.842 | 16.392 | 20.329 | 49.167 | 36.533 |
| 17 | 21.673 | 25.774 | 38.868 | 20.555 | 29.71 | 32.265 | 6.995 | 30.504 | 32.436 | 29.314 | 54.422 | 24.077 | 18.892 | 26.8 | 68.399 | 39.315 |
| 18 | 35.035 | 17.462 | 37.751 | 12.463 | 36.316 | 31.365 | 7.412 | 39.061 | 32.524 | 58.201 | 36.544 | 41.415 | 13.463 | 16.517 | 55.519 | 28.952 |
| 19 | 38.627 | 22.943 | 32.571 | 23.196 | 74.468 | 25.885 | 4.207 | 18.985 | 31.388 | 39.369 | 32.979 | 33.491 | 30.642 | 30.756 | 43.576 | 16.918 |
| 20 | 87.093 | 33.849 | 33.288 | 20.895 | 54.4 | 32.931 | 3.303 | 28.318 | 30.917 | 21.639 | 29.746 | 31.032 | 16.446 | 18.901 | 21.372 | 35.871 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.028 | -0.556 | 0.535 | -0.157 | -0.466 | -0.306 | -2.093 | 0.476 | 0.038 | -0.206 | 0.391 | 0.413 | -1.639 | -0.516 | 0.405 | 0.474 |
| 02 | 0.181 | -1.224 | 0.281 | -2.64 | 0.292 | -0.172 | -0.698 | -3.179 | 1.052 | -0.136 | -0.041 | -2.189 | 0.739 | 0.113 | 0.857 | -4.4 |
| 03 | -4.4 | -2.93 | -4.4 | 2.004 | -4.4 | -4.4 | -4.4 | 1.129 | -4.4 | -4.4 | -2.276 | 1.612 | -4.4 | -4.4 | -4.4 | -1.607 |
| 04 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.93 | -4.4 | -4.4 | -3.041 | -2.701 | -2.499 | -2.618 | 1.746 | 2.286 |
| 05 | -3.054 | -3.051 | -4.4 | -4.4 | -3.234 | -3.083 | -4.4 | -4.4 | 1.446 | -2.715 | -1.338 | 0.204 | 0.459 | -2.688 | 1.926 | 0.378 |
| 06 | -0.634 | 0.018 | -1.268 | 1.407 | -4.4 | -4.4 | -4.4 | -1.658 | 0.21 | 0.686 | -1.358 | 1.3 | -2.09 | -0.062 | -2.691 | 0.457 |
| 07 | -4.4 | -4.4 | 0.622 | -4.4 | -4.4 | -4.4 | 1.352 | -2.712 | -4.4 | -4.4 | -0.541 | -4.4 | -3.256 | -4.4 | 2.244 | -3.14 |
| 08 | -4.4 | -3.256 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 0.707 | 2.543 | -4.4 | -0.019 | -4.4 | -2.328 | -4.4 | -4.4 |
| 09 | 0.677 | -2.63 | -4.4 | -4.4 | 2.552 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -0.019 | -4.4 | -4.4 | -4.4 |
| 10 | 2.753 | -4.4 | -2.482 | -4.4 | -2.63 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 11 | -0.455 | 1.548 | 0.569 | 2.161 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.482 | -4.4 | -4.4 | -4.4 | -4.4 |
| 12 | -1.948 | -1.258 | -2.603 | -1.77 | 0.61 | 0.301 | -2.603 | 0.388 | 0.003 | -0.744 | -1.262 | -3.155 | 0.954 | 1.064 | 0.675 | 0.281 |
| 13 | -0.244 | 0.45 | 0.802 | 0.001 | 0.397 | 0.354 | -0.445 | 0.376 | -0.723 | -0.193 | -0.942 | -0.363 | -0.984 | 0.018 | -0.071 | -0.374 |
| 14 | -0.318 | -0.334 | -0.061 | -0.292 | 0.507 | -0.001 | -0.775 | 0.54 | -0.055 | 0.299 | -0.13 | 0.018 | -0.368 | 0.119 | -0.049 | 0.067 |
| 15 | -0.46 | 0.054 | 0.523 | -0.426 | 0.122 | 0.37 | -1.674 | 0.354 | -0.671 | 0.296 | -0.259 | -0.502 | -0.632 | 0.304 | 0.356 | 0.211 |
| 16 | -0.616 | -0.952 | 0.209 | -0.437 | 0.286 | 0.327 | -1.13 | 0.771 | 0.024 | -0.268 | 0.308 | -0.079 | -0.634 | -0.423 | 0.449 | 0.154 |
| 17 | -0.361 | -0.19 | 0.216 | -0.413 | -0.05 | 0.032 | -1.455 | -0.024 | 0.037 | -0.063 | 0.549 | -0.257 | -0.495 | -0.152 | 0.777 | 0.227 |
| 18 | 0.113 | -0.572 | 0.187 | -0.901 | 0.149 | 0.004 | -1.4 | 0.221 | 0.039 | 0.616 | 0.155 | 0.279 | -0.826 | -0.627 | 0.569 | -0.075 |
| 19 | 0.21 | -0.305 | 0.041 | -0.294 | 0.861 | -0.186 | -1.929 | -0.49 | 0.004 | 0.228 | 0.053 | 0.068 | -0.019 | -0.016 | 0.329 | -0.603 |
| 20 | 1.017 | 0.079 | 0.062 | -0.396 | 0.549 | 0.052 | -2.148 | -0.097 | -0.011 | -0.362 | -0.049 | -0.007 | -0.631 | -0.495 | -0.374 | 0.136 |