Model info
| Transcription factor | Nfya | ||||||||
| Model | NFYA_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 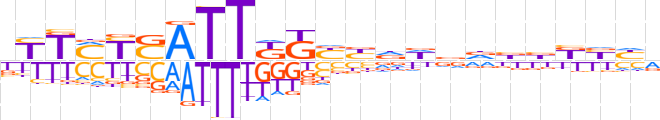 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Consensus | bdRMvvhhhhvRMMAATSRKARv | ||||||||
| wAUC | 0.8070458887767631 | ||||||||
| Best AUC | 0.8070458887767631 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 579 | ||||||||
| TF family | Heteromeric CCAAT-binding factors{4.2.1} | ||||||||
| TF subfamily | NF-YA (CP1A, CBF-B){4.2.1.0.1} | ||||||||
| MGI | 97316 | ||||||||
| EntrezGene | 18044 | ||||||||
| UniProt ID | NFYA_MOUSE | ||||||||
| UniProt AC | P23708 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.573 | 3.719 | 22.509 | 4.573 | 30.299 | 5.423 | 20.301 | 44.375 | 10.42 | 5.479 | 21.58 | 5.58 | 9.639 | 10.335 | 130.047 | 1.898 |
| 02 | 17.09 | 8.399 | 27.443 | 3.998 | 9.168 | 7.378 | 4.464 | 3.947 | 135.364 | 25.509 | 21.506 | 12.059 | 36.088 | 4.673 | 7.59 | 8.075 |
| 03 | 168.273 | 13.653 | 12.903 | 2.882 | 10.158 | 16.56 | 10.869 | 8.372 | 18.468 | 21.216 | 15.629 | 5.689 | 6.644 | 10.533 | 6.397 | 4.504 |
| 04 | 127.591 | 43.483 | 20.665 | 11.804 | 17.095 | 18.894 | 13.731 | 12.241 | 17.465 | 20.981 | 4.626 | 2.726 | 2.922 | 8.289 | 7.528 | 2.707 |
| 05 | 131.378 | 7.515 | 22.488 | 3.694 | 17.539 | 52.386 | 12.666 | 9.055 | 15.331 | 17.5 | 6.443 | 7.276 | 4.502 | 10.944 | 4.681 | 9.352 |
| 06 | 23.661 | 22.228 | 10.979 | 111.882 | 11.239 | 55.79 | 4.459 | 16.857 | 14.153 | 17.185 | 3.627 | 11.313 | 1.829 | 17.051 | 2.89 | 7.607 |
| 07 | 6.462 | 21.895 | 16.758 | 5.767 | 27.449 | 17.196 | 13.809 | 53.8 | 6.457 | 5.594 | 5.369 | 4.535 | 18.397 | 110.541 | 15.779 | 2.943 |
| 08 | 18.753 | 19.457 | 7.234 | 13.32 | 120.675 | 5.511 | 4.636 | 24.404 | 16.043 | 17.425 | 5.467 | 12.78 | 35.659 | 12.057 | 5.595 | 13.734 |
| 09 | 43.311 | 88.19 | 10.522 | 49.106 | 1.051 | 16.293 | 3.599 | 33.507 | 0.0 | 13.623 | 3.707 | 5.602 | 3.628 | 33.422 | 3.73 | 23.459 |
| 10 | 4.732 | 33.383 | 9.874 | 0.0 | 52.099 | 13.076 | 79.918 | 6.434 | 14.54 | 0.915 | 2.875 | 3.228 | 27.032 | 4.364 | 77.547 | 2.73 |
| 11 | 17.797 | 4.657 | 66.78 | 9.17 | 3.491 | 34.563 | 10.022 | 3.662 | 26.746 | 5.712 | 125.633 | 12.124 | 1.811 | 3.124 | 5.254 | 2.203 |
| 12 | 4.622 | 45.222 | 0.0 | 0.0 | 7.319 | 39.689 | 1.049 | 0.0 | 109.902 | 96.032 | 0.889 | 0.867 | 1.33 | 24.863 | 0.0 | 0.966 |
| 13 | 115.614 | 5.421 | 0.0 | 2.138 | 3.873 | 165.887 | 0.0 | 36.046 | 1.049 | 0.889 | 0.0 | 0.0 | 0.0 | 0.966 | 0.0 | 0.867 |
| 14 | 120.536 | 0.0 | 0.0 | 0.0 | 173.163 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 39.051 | 0.0 | 0.0 | 0.0 |
| 15 | 322.261 | 0.868 | 6.677 | 2.944 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 2.693 | 32.511 | 6.429 | 280.628 | 0.0 | 0.0 | 0.0 | 0.868 | 0.875 | 0.928 | 0.958 | 3.915 | 0.0 | 0.0 | 0.948 | 1.996 |
| 17 | 0.0 | 0.875 | 2.693 | 0.0 | 0.0 | 33.439 | 0.0 | 0.0 | 0.0 | 3.612 | 4.724 | 0.0 | 7.428 | 92.044 | 184.115 | 3.82 |
| 18 | 1.834 | 0.0 | 5.594 | 0.0 | 72.956 | 34.966 | 20.153 | 1.894 | 163.811 | 0.898 | 25.778 | 1.046 | 2.028 | 0.0 | 1.792 | 0.0 |
| 19 | 14.9 | 15.801 | 193.637 | 16.29 | 0.898 | 2.604 | 0.875 | 31.487 | 5.522 | 11.15 | 35.748 | 0.898 | 0.0 | 0.0 | 2.94 | 0.0 |
| 20 | 14.989 | 0.866 | 3.665 | 1.8 | 17.579 | 0.968 | 10.084 | 0.924 | 192.401 | 24.013 | 13.918 | 2.87 | 41.274 | 5.533 | 0.879 | 0.988 |
| 21 | 203.536 | 2.77 | 49.936 | 10.0 | 9.149 | 5.859 | 9.21 | 7.162 | 16.344 | 0.942 | 9.405 | 1.855 | 1.827 | 0.0 | 3.767 | 0.988 |
| 22 | 139.635 | 48.63 | 32.794 | 9.796 | 3.76 | 4.946 | 0.0 | 0.866 | 13.916 | 34.646 | 17.516 | 6.24 | 0.849 | 2.804 | 10.858 | 5.493 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.115 | -1.645 | 0.078 | -1.456 | 0.371 | -1.297 | -0.024 | 0.749 | -0.674 | -1.287 | 0.036 | -1.27 | -0.749 | -0.682 | 1.819 | -2.236 |
| 02 | -0.193 | -0.882 | 0.273 | -1.579 | -0.798 | -1.006 | -1.478 | -1.591 | 1.859 | 0.201 | 0.033 | -0.533 | 0.544 | -1.435 | -0.979 | -0.919 |
| 03 | 2.076 | -0.412 | -0.467 | -1.875 | -0.699 | -0.223 | -0.633 | -0.885 | -0.117 | 0.02 | -0.28 | -1.252 | -1.105 | -0.664 | -1.141 | -1.47 |
| 04 | 1.8 | 0.729 | -0.006 | -0.553 | -0.192 | -0.094 | -0.406 | -0.518 | -0.171 | 0.009 | -1.445 | -1.924 | -1.863 | -0.894 | -0.986 | -1.93 |
| 05 | 1.829 | -0.988 | 0.077 | -1.652 | -0.167 | 0.913 | -0.485 | -0.81 | -0.299 | -0.169 | -1.134 | -1.019 | -1.47 | -0.627 | -1.434 | -0.778 |
| 06 | 0.127 | 0.065 | -0.624 | 1.669 | -0.601 | 0.976 | -1.479 | -0.206 | -0.377 | -0.187 | -1.668 | -0.595 | -2.267 | -0.195 | -1.873 | -0.976 |
| 07 | -1.132 | 0.051 | -0.212 | -1.239 | 0.273 | -0.187 | -0.401 | 0.94 | -1.132 | -1.268 | -1.306 | -1.463 | -0.12 | 1.657 | -0.271 | -1.856 |
| 08 | -0.102 | -0.065 | -1.024 | -0.436 | 1.744 | -1.282 | -1.443 | 0.157 | -0.254 | -0.174 | -1.289 | -0.476 | 0.532 | -0.533 | -1.267 | -0.406 |
| 09 | 0.725 | 1.432 | -0.665 | 0.849 | -2.706 | -0.239 | -1.675 | 0.47 | -4.066 | -0.414 | -1.648 | -1.266 | -1.668 | 0.468 | -1.643 | 0.119 |
| 10 | -1.424 | 0.467 | -0.726 | -4.066 | 0.908 | -0.454 | 1.333 | -1.136 | -0.351 | -2.807 | -1.877 | -1.774 | 0.258 | -1.499 | 1.303 | -1.923 |
| 11 | -0.153 | -1.439 | 1.155 | -0.797 | -1.703 | 0.501 | -0.712 | -1.66 | 0.248 | -1.248 | 1.784 | -0.527 | -2.276 | -1.803 | -1.326 | -2.11 |
| 12 | -1.446 | 0.767 | -4.066 | -4.066 | -1.013 | 0.638 | -2.707 | -4.066 | 1.651 | 1.516 | -2.827 | -2.845 | -2.526 | 0.176 | -4.066 | -2.768 |
| 13 | 1.701 | -1.297 | -4.066 | -2.135 | -1.609 | 2.061 | -4.066 | 0.543 | -2.707 | -2.827 | -4.066 | -4.066 | -4.066 | -2.768 | -4.066 | -2.845 |
| 14 | 1.743 | -4.066 | -4.066 | -4.066 | 2.104 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | 0.622 | -4.066 | -4.066 | -4.066 |
| 15 | 2.724 | -2.845 | -1.1 | -1.856 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 | -4.066 |
| 16 | -1.935 | 0.441 | -1.136 | 2.586 | -4.066 | -4.066 | -4.066 | -2.845 | -2.839 | -2.796 | -2.774 | -1.599 | -4.066 | -4.066 | -2.781 | -2.194 |
| 17 | -4.066 | -2.839 | -1.935 | -4.066 | -4.066 | 0.468 | -4.066 | -4.066 | -4.066 | -1.672 | -1.425 | -4.066 | -0.999 | 1.474 | 2.165 | -1.621 |
| 18 | -2.265 | -4.066 | -1.267 | -4.066 | 1.243 | 0.513 | -0.031 | -2.238 | 2.049 | -2.821 | 0.211 | -2.709 | -2.18 | -4.066 | -2.284 | -4.066 |
| 19 | -0.327 | -0.269 | 2.216 | -0.24 | -2.821 | -1.965 | -2.839 | 0.409 | -1.28 | -0.609 | 0.534 | -2.821 | -4.066 | -4.066 | -1.857 | -4.066 |
| 20 | -0.321 | -2.846 | -1.659 | -2.28 | -0.165 | -2.766 | -0.706 | -2.8 | 2.209 | 0.141 | -0.393 | -1.879 | 0.677 | -1.278 | -2.835 | -2.752 |
| 21 | 2.266 | -1.91 | 0.866 | -0.714 | -0.8 | -1.224 | -0.793 | -1.034 | -0.236 | -2.786 | -0.773 | -2.256 | -2.268 | -4.066 | -1.634 | -2.752 |
| 22 | 1.89 | 0.84 | 0.449 | -0.734 | -1.636 | -1.383 | -4.066 | -2.846 | -0.393 | 0.504 | -0.168 | -1.165 | -2.86 | -1.899 | -0.634 | -1.285 |