Model info
| Transcription factor | Stat5b | ||||||||
| Model | STA5B_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 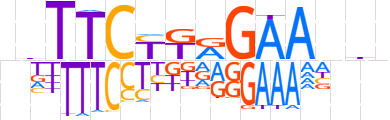 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Consensus | ndTTCYTRGAAnhn | ||||||||
| wAUC | 0.9507193214364849 | ||||||||
| Best AUC | 0.9507193214364849 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 503 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT5B{6.2.1.0.6} | ||||||||
| MGI | 103035 | ||||||||
| EntrezGene | 20851 | ||||||||
| UniProt ID | STA5B_MOUSE | ||||||||
| UniProt AC | P42232 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 44.592 | 23.873 | 48.258 | 61.442 | 23.406 | 16.738 | 6.177 | 40.873 | 28.779 | 23.418 | 28.22 | 42.14 | 5.863 | 17.175 | 27.958 | 61.088 |
| 02 | 0.0 | 0.944 | 0.0 | 101.696 | 0.0 | 0.0 | 0.976 | 80.229 | 0.0 | 1.066 | 0.0 | 109.547 | 0.0 | 0.0 | 0.0 | 205.543 |
| 03 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.066 | 0.0 | 0.944 | 0.0 | 0.0 | 0.0 | 0.976 | 24.093 | 4.055 | 0.0 | 468.866 |
| 04 | 0.0 | 23.1 | 0.0 | 0.993 | 0.941 | 4.18 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.268 | 461.783 | 0.0 | 2.735 |
| 05 | 0.0 | 7.21 | 0.0 | 0.0 | 20.509 | 134.396 | 4.959 | 329.199 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.837 | 0.0 | 0.89 |
| 06 | 0.0 | 0.0 | 13.888 | 6.621 | 0.0 | 0.0 | 8.518 | 135.924 | 0.0 | 0.0 | 2.948 | 2.01 | 0.0 | 0.0 | 204.63 | 125.46 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 160.62 | 1.978 | 55.097 | 12.289 | 102.029 | 3.033 | 156.916 | 8.038 |
| 08 | 4.063 | 1.816 | 254.929 | 1.841 | 0.0 | 0.0 | 5.011 | 0.0 | 2.894 | 0.0 | 208.205 | 0.914 | 0.0 | 1.065 | 19.263 | 0.0 |
| 09 | 6.957 | 0.0 | 0.0 | 0.0 | 1.995 | 0.0 | 0.0 | 0.886 | 438.056 | 3.867 | 0.942 | 44.542 | 2.755 | 0.0 | 0.0 | 0.0 |
| 10 | 438.245 | 2.943 | 7.576 | 0.999 | 3.867 | 0.0 | 0.0 | 0.0 | 0.942 | 0.0 | 0.0 | 0.0 | 42.662 | 1.871 | 0.894 | 0.0 |
| 11 | 178.33 | 110.544 | 85.313 | 111.53 | 1.958 | 0.0 | 0.958 | 1.898 | 3.771 | 1.831 | 1.86 | 1.008 | 0.0 | 0.0 | 0.0 | 0.999 |
| 12 | 47.884 | 40.707 | 27.108 | 68.361 | 26.084 | 17.275 | 1.876 | 67.141 | 17.962 | 28.065 | 10.798 | 31.305 | 17.923 | 24.365 | 22.388 | 50.759 |
| 13 | 25.891 | 27.609 | 34.489 | 21.865 | 41.523 | 33.996 | 4.86 | 30.033 | 20.379 | 14.697 | 16.925 | 10.167 | 28.128 | 58.759 | 87.473 | 43.206 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.352 | -0.265 | 0.43 | 0.67 | -0.285 | -0.614 | -1.573 | 0.266 | -0.081 | -0.284 | -0.101 | 0.296 | -1.622 | -0.589 | -0.11 | 0.664 |
| 02 | -4.4 | -3.167 | -4.4 | 1.171 | -4.4 | -4.4 | -3.144 | 0.935 | -4.4 | -3.08 | -4.4 | 1.246 | -4.4 | -4.4 | -4.4 | 1.873 |
| 03 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.08 | -4.4 | -3.167 | -4.4 | -4.4 | -4.4 | -3.144 | -0.256 | -1.963 | -4.4 | 2.697 |
| 04 | -4.4 | -0.298 | -4.4 | -3.131 | -3.169 | -1.935 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.559 | 2.682 | -4.4 | -2.316 |
| 05 | -4.4 | -1.426 | -4.4 | -4.4 | -0.415 | 1.449 | -1.778 | 2.343 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.283 | -4.4 | -3.209 |
| 06 | -4.4 | -4.4 | -0.796 | -1.507 | -4.4 | -4.4 | -1.268 | 1.461 | -4.4 | -4.4 | -2.249 | -2.579 | -4.4 | -4.4 | 1.869 | 1.381 |
| 07 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 1.627 | -2.593 | 0.562 | -0.915 | 1.175 | -2.224 | 1.604 | -1.323 |
| 08 | -1.961 | -2.664 | 2.088 | -2.653 | -4.4 | -4.4 | -1.768 | -4.4 | -2.266 | -4.4 | 1.886 | -3.19 | -4.4 | -3.081 | -0.476 | -4.4 |
| 09 | -1.46 | -4.4 | -4.4 | -4.4 | -2.586 | -4.4 | -4.4 | -3.212 | 2.629 | -2.006 | -3.169 | 0.351 | -2.309 | -4.4 | -4.4 | -4.4 |
| 10 | 2.629 | -2.251 | -1.379 | -3.127 | -2.006 | -4.4 | -4.4 | -4.4 | -3.169 | -4.4 | -4.4 | -4.4 | 0.308 | -2.639 | -3.205 | -4.4 |
| 11 | 1.731 | 1.255 | 0.996 | 1.263 | -2.601 | -4.4 | -3.157 | -2.627 | -2.029 | -2.657 | -2.644 | -3.121 | -4.4 | -4.4 | -4.4 | -3.127 |
| 12 | 0.422 | 0.262 | -0.14 | 0.776 | -0.178 | -0.583 | -2.637 | 0.758 | -0.545 | -0.106 | -1.04 | 0.002 | -0.547 | -0.245 | -0.329 | 0.48 |
| 13 | -0.186 | -0.122 | 0.097 | -0.352 | 0.281 | 0.083 | -1.796 | -0.039 | -0.421 | -0.741 | -0.603 | -1.098 | -0.104 | 0.626 | 1.021 | 0.321 |