Model info
| Transcription factor | VDR | ||||||||
| Model | VDR_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 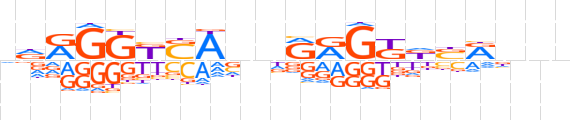 | ||||||||
| LOGO (reverse complement) | 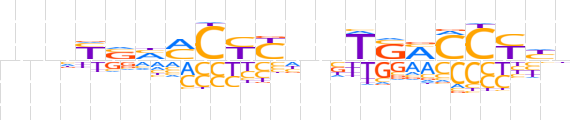 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | B | ||||||||
| Consensus | ndRGGKSAbnRRGKdSRnnn | ||||||||
| wAUC | 0.8065594918260511 | ||||||||
| Best AUC | 0.9047642709441346 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 508 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | Vitamin D receptor (NR1I){2.1.2.4} | ||||||||
| HGNC | 12679 | ||||||||
| EntrezGene | 7421 | ||||||||
| UniProt ID | VDR_HUMAN | ||||||||
| UniProt AC | P11473 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 49.163 | 6.096 | 88.341 | 13.332 | 59.794 | 16.702 | 14.615 | 16.614 | 54.796 | 8.809 | 59.707 | 11.207 | 24.259 | 4.587 | 62.12 | 9.858 |
| 02 | 96.284 | 1.856 | 86.208 | 3.665 | 24.22 | 1.876 | 9.179 | 0.918 | 117.281 | 0.969 | 106.532 | 0.0 | 17.72 | 0.878 | 30.524 | 1.888 |
| 03 | 35.336 | 1.303 | 206.605 | 12.261 | 4.542 | 0.0 | 1.038 | 0.0 | 20.84 | 0.0 | 204.115 | 7.488 | 0.0 | 0.0 | 6.471 | 0.0 |
| 04 | 3.992 | 2.819 | 51.043 | 2.865 | 1.303 | 0.0 | 0.0 | 0.0 | 6.674 | 25.392 | 331.422 | 54.741 | 1.837 | 0.0 | 14.832 | 3.081 |
| 05 | 0.0 | 0.0 | 2.95 | 10.856 | 1.008 | 1.871 | 2.685 | 22.647 | 21.283 | 46.58 | 75.878 | 253.557 | 2.726 | 5.58 | 6.359 | 46.022 |
| 06 | 3.58 | 16.806 | 4.631 | 0.0 | 4.642 | 40.417 | 1.963 | 7.009 | 2.712 | 55.807 | 20.923 | 8.43 | 3.073 | 233.844 | 66.307 | 29.857 |
| 07 | 11.22 | 0.0 | 0.0 | 2.786 | 297.715 | 7.82 | 14.245 | 27.095 | 66.099 | 1.841 | 16.143 | 9.741 | 29.902 | 1.913 | 9.255 | 4.226 |
| 08 | 71.596 | 128.841 | 145.031 | 59.468 | 2.86 | 4.695 | 0.0 | 4.018 | 2.717 | 20.589 | 5.043 | 11.294 | 2.789 | 10.008 | 24.052 | 6.999 |
| 09 | 15.457 | 13.522 | 23.199 | 27.784 | 61.549 | 24.187 | 13.484 | 64.914 | 41.008 | 32.609 | 63.557 | 36.953 | 14.118 | 20.114 | 20.626 | 26.92 |
| 10 | 23.85 | 12.425 | 94.856 | 1.001 | 48.509 | 12.431 | 25.299 | 4.192 | 13.243 | 13.239 | 91.553 | 2.831 | 12.757 | 0.923 | 139.359 | 3.532 |
| 11 | 39.726 | 4.144 | 52.481 | 2.009 | 31.564 | 0.852 | 4.714 | 1.888 | 182.632 | 6.936 | 155.749 | 5.75 | 4.694 | 0.923 | 5.939 | 0.0 |
| 12 | 13.825 | 3.781 | 234.269 | 6.742 | 4.939 | 0.0 | 7.917 | 0.0 | 15.141 | 6.088 | 185.107 | 12.548 | 1.797 | 0.0 | 7.849 | 0.0 |
| 13 | 1.833 | 0.872 | 28.634 | 4.363 | 0.0 | 0.0 | 1.856 | 8.013 | 10.222 | 10.085 | 201.869 | 212.965 | 0.0 | 3.836 | 8.841 | 6.612 |
| 14 | 3.984 | 0.859 | 2.75 | 4.461 | 1.766 | 0.859 | 0.831 | 11.338 | 49.724 | 18.156 | 74.902 | 98.418 | 19.5 | 11.712 | 45.298 | 155.442 |
| 15 | 10.007 | 33.549 | 26.795 | 4.623 | 3.139 | 20.492 | 2.334 | 5.622 | 13.589 | 70.672 | 29.043 | 10.477 | 4.461 | 163.418 | 51.077 | 50.702 |
| 16 | 13.86 | 4.938 | 9.66 | 2.738 | 217.092 | 15.872 | 29.54 | 25.627 | 65.654 | 7.769 | 30.346 | 5.481 | 31.336 | 5.976 | 30.234 | 3.879 |
| 17 | 73.857 | 69.188 | 95.147 | 89.75 | 13.777 | 7.947 | 0.961 | 11.87 | 16.248 | 18.359 | 27.939 | 37.234 | 6.616 | 13.563 | 6.584 | 10.961 |
| 18 | 31.867 | 10.89 | 57.466 | 10.275 | 39.889 | 23.061 | 15.165 | 30.942 | 37.173 | 23.213 | 44.543 | 25.701 | 16.735 | 23.295 | 66.037 | 43.747 |
| 19 | 22.38 | 27.473 | 49.154 | 26.657 | 14.875 | 39.438 | 6.25 | 19.897 | 43.179 | 35.631 | 70.529 | 33.871 | 8.0 | 30.605 | 42.347 | 29.714 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.449 | -1.585 | 1.031 | -0.835 | 0.643 | -0.616 | -0.746 | -0.621 | 0.556 | -1.235 | 0.642 | -1.004 | -0.25 | -1.85 | 0.681 | -1.127 |
| 02 | 1.117 | -2.646 | 1.007 | -2.055 | -0.251 | -2.637 | -1.196 | -3.187 | 1.314 | -3.149 | 1.218 | -4.4 | -0.558 | -3.218 | -0.023 | -2.632 |
| 03 | 0.121 | -2.929 | 1.878 | -0.917 | -1.859 | -4.4 | -3.099 | -4.4 | -0.399 | -4.4 | 1.866 | -1.39 | -4.4 | -4.4 | -1.529 | -4.4 |
| 04 | -1.977 | -2.289 | 0.486 | -2.275 | -2.929 | -4.4 | -4.4 | -4.4 | -1.5 | -0.205 | 2.35 | 0.555 | -2.654 | -4.4 | -0.732 | -2.211 |
| 05 | -4.4 | -4.4 | -2.249 | -1.034 | -3.12 | -2.639 | -2.332 | -0.317 | -0.378 | 0.395 | 0.88 | 2.083 | -2.318 | -1.668 | -1.545 | 0.383 |
| 06 | -2.076 | -0.61 | -1.841 | -4.4 | -1.839 | 0.254 | -2.6 | -1.453 | -2.323 | 0.574 | -0.395 | -1.278 | -2.213 | 2.002 | 0.746 | -0.045 |
| 07 | -1.003 | -4.4 | -4.4 | -2.299 | 2.243 | -1.349 | -0.771 | -0.141 | 0.743 | -2.653 | -0.649 | -1.139 | -0.044 | -2.621 | -1.188 | -1.925 |
| 08 | 0.822 | 1.407 | 1.525 | 0.638 | -2.276 | -1.828 | -4.4 | -1.971 | -2.321 | -0.411 | -1.762 | -0.996 | -2.298 | -1.113 | -0.258 | -1.455 |
| 09 | -0.691 | -0.822 | -0.294 | -0.116 | 0.672 | -0.253 | -0.824 | 0.725 | 0.269 | 0.042 | 0.704 | 0.166 | -0.78 | -0.434 | -0.409 | -0.147 |
| 10 | -0.266 | -0.904 | 1.102 | -3.126 | 0.435 | -0.903 | -0.208 | -1.932 | -0.842 | -0.842 | 1.067 | -2.285 | -0.878 | -3.183 | 1.485 | -2.088 |
| 11 | 0.237 | -1.943 | 0.513 | -2.58 | 0.01 | -3.239 | -1.825 | -2.632 | 1.755 | -1.463 | 1.596 | -1.64 | -1.829 | -3.183 | -1.609 | -4.4 |
| 12 | -0.8 | -2.027 | 2.004 | -1.49 | -1.782 | -4.4 | -1.338 | -4.4 | -0.712 | -1.586 | 1.769 | -0.894 | -2.673 | -4.4 | -1.346 | -4.4 |
| 13 | -2.656 | -3.223 | -0.086 | -1.896 | -4.4 | -4.4 | -2.646 | -1.326 | -1.093 | -1.106 | 1.855 | 1.909 | -4.4 | -2.013 | -1.232 | -1.508 |
| 14 | -1.979 | -3.233 | -2.311 | -1.875 | -2.687 | -3.233 | -3.256 | -0.993 | 0.46 | -0.534 | 0.867 | 1.139 | -0.464 | -0.961 | 0.367 | 1.594 |
| 15 | -1.113 | 0.07 | -0.152 | -1.843 | -2.194 | -0.416 | -2.453 | -1.661 | -0.817 | 0.809 | -0.072 | -1.069 | -1.876 | 1.644 | 0.487 | 0.479 |
| 16 | -0.798 | -1.782 | -1.147 | -2.315 | 1.928 | -0.666 | -0.056 | -0.196 | 0.736 | -1.355 | -0.029 | -1.685 | 0.003 | -1.604 | -0.033 | -2.003 |
| 17 | 0.853 | 0.788 | 1.105 | 1.047 | -0.804 | -1.334 | -3.155 | -0.948 | -0.643 | -0.523 | -0.111 | 0.173 | -1.508 | -0.819 | -1.512 | -1.025 |
| 18 | 0.019 | -1.031 | 0.604 | -1.088 | 0.241 | -0.3 | -0.71 | -0.01 | 0.172 | -0.293 | 0.351 | -0.193 | -0.614 | -0.29 | 0.742 | 0.333 |
| 19 | -0.329 | -0.127 | 0.448 | -0.157 | -0.729 | 0.23 | -1.561 | -0.444 | 0.32 | 0.13 | 0.807 | 0.08 | -1.328 | -0.021 | 0.301 | -0.05 |