Model info
| Transcription factor | TFE3 | ||||||||
| Model | TFE3_HUMAN.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 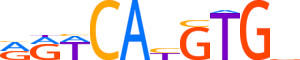 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 10 | ||||||||
| Quality | C | ||||||||
| Consensus | RRWCAYGTGv | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 17 | ||||||||
| TF family | bHLH-ZIP factors{1.2.6} | ||||||||
| TF subfamily | TFE3-like factors{1.2.6.1} | ||||||||
| HGNC | 11752 | ||||||||
| EntrezGene | 7030 | ||||||||
| UniProt ID | TFE3_HUMAN | ||||||||
| UniProt AC | P19532 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 7.65 | 1.102 | 8.028 | 0.22 |
| 02 | 4.785 | 0.0 | 11.081 | 1.133 |
| 03 | 4.156 | 1.133 | 0.0 | 11.711 |
| 04 | 0.0 | 17.0 | 0.0 | 0.0 |
| 05 | 17.0 | 0.0 | 0.0 | 0.0 |
| 06 | 1.133 | 6.548 | 1.133 | 8.185 |
| 07 | 0.0 | 2.896 | 14.104 | 0.0 |
| 08 | 1.133 | 0.0 | 0.0 | 15.867 |
| 09 | 0.0 | 0.0 | 17.0 | 0.0 |
| 10 | 3.904 | 3.778 | 6.8 | 2.519 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.522 | -1.008 | 0.566 | -1.675 |
| 02 | 0.102 | -1.946 | 0.866 | -0.99 |
| 03 | -0.019 | -0.99 | -1.946 | 0.918 |
| 04 | -1.946 | 1.273 | -1.946 | -1.946 |
| 05 | 1.273 | -1.946 | -1.946 | -1.946 |
| 06 | -0.99 | 0.381 | -0.99 | 0.584 |
| 07 | -1.946 | -0.319 | 1.094 | -1.946 |
| 08 | -0.99 | -1.946 | -1.946 | 1.207 |
| 09 | -1.946 | -1.946 | 1.273 | -1.946 |
| 10 | -0.072 | -0.1 | 0.415 | -0.43 |