Model info
| Transcription factor | TBX4 | ||||||||
| Model | TBX4_HUMAN.H10MO.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 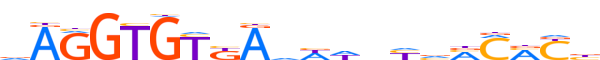 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HT-SELEX | ||||||||
| Model length | 20 | ||||||||
| Quality | D | ||||||||
| Consensus | dAGGTGYKAhWhnYvMMMYb | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 16745 | ||||||||
| TF family | TBX2-related factors{6.5.4} | ||||||||
| TF subfamily | TBX4{6.5.4.0.3} | ||||||||
| HGNC | 11603 | ||||||||
| EntrezGene | 9496 | ||||||||
| UniProt ID | TBX4_HUMAN | ||||||||
| UniProt AC | P57082 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 6793.75 | 1707.75 | 4266.75 | 2945.75 |
| 02 | 13760.75 | 522.75 | 802.75 | 627.75 |
| 03 | 1115.5 | 1036.5 | 13049.5 | 512.5 |
| 04 | 245.75 | 269.75 | 14955.75 | 242.75 |
| 05 | 395.25 | 1164.25 | 397.25 | 13757.25 |
| 06 | 374.5 | 352.5 | 14634.5 | 352.5 |
| 07 | 1236.75 | 1579.75 | 462.75 | 12434.75 |
| 08 | 733.5 | 2494.5 | 9714.5 | 2771.5 |
| 09 | 13562.0 | 451.0 | 1197.0 | 504.0 |
| 10 | 6727.0 | 2759.0 | 2453.0 | 3775.0 |
| 11 | 9033.25 | 1332.25 | 1336.25 | 4012.25 |
| 12 | 5468.25 | 1452.25 | 1214.25 | 7579.25 |
| 13 | 3916.75 | 3466.75 | 3430.75 | 4899.75 |
| 14 | 1926.0 | 3481.0 | 1142.0 | 9165.0 |
| 15 | 4874.25 | 5834.25 | 3875.25 | 1130.25 |
| 16 | 9696.75 | 2116.75 | 2075.75 | 1824.75 |
| 17 | 1748.25 | 12343.25 | 873.25 | 749.25 |
| 18 | 10175.0 | 2299.0 | 2260.0 | 980.0 |
| 19 | 1060.0 | 12011.0 | 1305.0 | 1338.0 |
| 20 | 1783.75 | 7580.75 | 2594.75 | 3754.75 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.547 | -0.832 | 0.083 | -0.288 |
| 02 | 1.253 | -2.013 | -1.586 | -1.831 |
| 03 | -1.257 | -1.331 | 1.2 | -2.033 |
| 04 | -2.763 | -2.67 | 1.336 | -2.775 |
| 05 | -2.291 | -1.215 | -2.286 | 1.253 |
| 06 | -2.345 | -2.405 | 1.315 | -2.405 |
| 07 | -1.154 | -0.91 | -2.134 | 1.152 |
| 08 | -1.676 | -0.454 | 0.905 | -0.349 |
| 09 | 1.239 | -2.16 | -1.187 | -2.049 |
| 10 | 0.538 | -0.353 | -0.471 | -0.04 |
| 11 | 0.832 | -1.08 | -1.077 | 0.021 |
| 12 | 0.331 | -0.994 | -1.173 | 0.657 |
| 13 | -0.003 | -0.125 | -0.135 | 0.221 |
| 14 | -0.712 | -0.121 | -1.234 | 0.847 |
| 15 | 0.216 | 0.395 | -0.014 | -1.244 |
| 16 | 0.903 | -0.618 | -0.637 | -0.766 |
| 17 | -0.809 | 1.144 | -1.502 | -1.654 |
| 18 | 0.951 | -0.535 | -0.552 | -1.387 |
| 19 | -1.308 | 1.117 | -1.101 | -1.076 |
| 20 | -0.789 | 0.657 | -0.414 | -0.045 |