Model info
| Transcription factor | STAT4 | ||||||||
| Model | STAT4_HUMAN.H10DI.D | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 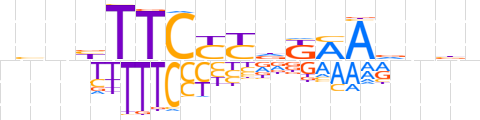 | ||||||||
| LOGO (reverse complement) | 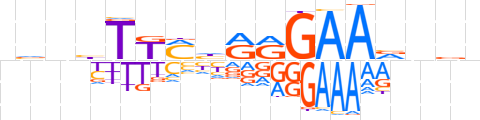 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 17 | ||||||||
| Quality | D | ||||||||
| Consensus | nbnbTTCYYdKMAvnbn | ||||||||
| wAUC | 0.7940708803638311 | ||||||||
| Best AUC | 0.7940708803638311 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 493 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT4{6.2.1.0.4} | ||||||||
| HGNC | 11365 | ||||||||
| EntrezGene | 6775 | ||||||||
| UniProt ID | STAT4_HUMAN | ||||||||
| UniProt AC | Q14765 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 10.79 | 21.344 | 24.487 | 20.643 | 32.44 | 84.756 | 10.698 | 47.391 | 7.829 | 47.892 | 31.177 | 17.876 | 7.126 | 49.83 | 47.231 | 29.489 |
| 02 | 17.869 | 11.855 | 23.774 | 4.688 | 85.957 | 50.758 | 15.721 | 51.385 | 23.185 | 29.166 | 49.687 | 11.556 | 16.15 | 31.215 | 39.826 | 28.209 |
| 03 | 14.743 | 31.836 | 23.987 | 72.594 | 15.384 | 39.481 | 5.958 | 62.171 | 10.636 | 37.505 | 14.501 | 66.367 | 6.638 | 24.735 | 10.129 | 54.336 |
| 04 | 0.0 | 1.889 | 0.0 | 45.513 | 0.0 | 4.928 | 0.0 | 128.629 | 0.0 | 0.0 | 0.0 | 54.574 | 0.0 | 3.14 | 0.972 | 251.356 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.957 | 0.0 | 0.0 | 0.0 | 0.972 | 1.921 | 0.0 | 23.865 | 454.286 |
| 06 | 0.0 | 1.921 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 22.894 | 0.0 | 0.971 | 16.995 | 439.266 | 2.031 | 6.923 |
| 07 | 3.18 | 10.649 | 0.0 | 3.166 | 13.307 | 268.004 | 2.185 | 180.584 | 0.0 | 1.04 | 0.0 | 0.992 | 0.0 | 7.893 | 0.0 | 0.0 |
| 08 | 0.0 | 15.447 | 0.0 | 1.04 | 0.0 | 136.919 | 4.061 | 146.606 | 0.0 | 0.0 | 0.0 | 2.185 | 0.0 | 114.662 | 6.143 | 63.937 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 125.203 | 29.246 | 72.796 | 39.783 | 0.0 | 4.171 | 6.032 | 0.0 | 24.544 | 14.439 | 140.351 | 34.435 |
| 10 | 1.89 | 9.112 | 125.058 | 13.688 | 9.753 | 4.156 | 15.502 | 18.444 | 10.121 | 9.781 | 139.402 | 59.874 | 2.923 | 2.984 | 49.479 | 18.833 |
| 11 | 14.925 | 8.798 | 0.0 | 0.964 | 12.283 | 13.751 | 0.0 | 0.0 | 241.792 | 66.343 | 6.361 | 14.945 | 68.922 | 36.244 | 3.727 | 1.946 |
| 12 | 317.572 | 3.109 | 11.891 | 5.351 | 104.032 | 11.568 | 0.0 | 9.535 | 7.986 | 1.123 | 0.979 | 0.0 | 13.047 | 2.895 | 1.019 | 0.893 |
| 13 | 177.308 | 58.019 | 158.709 | 48.601 | 6.538 | 3.127 | 2.944 | 6.087 | 0.0 | 5.049 | 6.101 | 2.74 | 1.915 | 3.852 | 3.204 | 6.807 |
| 14 | 46.241 | 53.399 | 52.988 | 33.133 | 24.541 | 13.872 | 4.32 | 27.315 | 45.24 | 51.503 | 47.645 | 26.57 | 4.876 | 17.56 | 26.276 | 15.522 |
| 15 | 12.904 | 32.597 | 54.949 | 20.448 | 8.956 | 60.427 | 13.282 | 53.669 | 11.592 | 39.171 | 32.561 | 47.905 | 3.785 | 24.31 | 57.729 | 16.716 |
| 16 | 7.695 | 6.853 | 15.831 | 6.857 | 50.351 | 48.535 | 19.119 | 38.499 | 30.928 | 45.318 | 55.528 | 26.747 | 12.698 | 23.48 | 67.263 | 35.297 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.022 | -0.358 | -0.223 | -0.39 | 0.055 | 1.008 | -1.031 | 0.43 | -1.33 | 0.441 | 0.016 | -0.532 | -1.42 | 0.48 | 0.427 | -0.039 |
| 02 | -0.532 | -0.932 | -0.252 | -1.812 | 1.022 | 0.498 | -0.657 | 0.51 | -0.276 | -0.05 | 0.477 | -0.956 | -0.631 | 0.017 | 0.258 | -0.083 |
| 03 | -0.72 | 0.036 | -0.243 | 0.854 | -0.678 | 0.249 | -1.589 | 0.7 | -1.036 | 0.198 | -0.736 | 0.765 | -1.487 | -0.213 | -1.084 | 0.566 |
| 04 | -4.385 | -2.614 | -4.385 | 0.39 | -4.385 | -1.766 | -4.385 | 1.424 | -4.385 | -4.385 | -4.385 | 0.57 | -4.385 | -2.176 | -3.13 | 2.092 |
| 05 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -1.1 | -4.385 | -4.385 | -4.385 | -3.13 | -2.6 | -4.385 | -0.248 | 2.683 |
| 06 | -4.385 | -2.6 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -4.385 | -0.289 | -4.385 | -3.131 | -0.581 | 2.65 | -2.553 | -1.447 |
| 07 | -2.165 | -1.035 | -4.385 | -2.169 | -0.819 | 2.156 | -2.492 | 1.762 | -4.385 | -3.081 | -4.385 | -3.115 | -4.385 | -1.322 | -4.385 | -4.385 |
| 08 | -4.385 | -0.674 | -4.385 | -3.08 | -4.385 | 1.486 | -1.944 | 1.554 | -4.385 | -4.385 | -4.385 | -2.492 | -4.385 | 1.309 | -1.56 | 0.728 |
| 09 | -4.385 | -4.385 | -4.385 | -4.385 | 1.397 | -0.048 | 0.857 | 0.257 | -4.385 | -1.919 | -1.577 | -4.385 | -0.22 | -0.74 | 1.511 | 0.114 |
| 10 | -2.614 | -1.185 | 1.395 | -0.792 | -1.12 | -1.923 | -0.671 | -0.501 | -1.084 | -1.117 | 1.504 | 0.662 | -2.24 | -2.221 | 0.473 | -0.48 |
| 11 | -0.708 | -1.219 | -4.385 | -3.136 | -0.897 | -0.788 | -4.385 | -4.385 | 2.053 | 0.764 | -1.527 | -0.706 | 0.802 | 0.165 | -2.022 | -2.589 |
| 12 | 2.326 | -2.185 | -0.929 | -1.689 | 1.212 | -0.955 | -4.385 | -1.142 | -1.311 | -3.024 | -3.124 | -4.385 | -0.839 | -2.248 | -3.095 | -3.189 |
| 13 | 1.744 | 0.631 | 1.633 | 0.455 | -1.501 | -2.18 | -2.233 | -1.569 | -4.385 | -1.743 | -1.566 | -2.296 | -2.602 | -1.992 | -2.158 | -1.463 |
| 14 | 0.406 | 0.549 | 0.541 | 0.076 | -0.22 | -0.779 | -1.887 | -0.115 | 0.384 | 0.513 | 0.435 | -0.142 | -1.776 | -0.549 | -0.153 | -0.669 |
| 15 | -0.849 | 0.06 | 0.577 | -0.4 | -1.202 | 0.671 | -0.821 | 0.554 | -0.953 | 0.241 | 0.059 | 0.441 | -2.008 | -0.23 | 0.626 | -0.597 |
| 16 | -1.347 | -1.457 | -0.65 | -1.456 | 0.49 | 0.454 | -0.466 | 0.224 | 0.008 | 0.386 | 0.587 | -0.136 | -0.865 | -0.264 | 0.778 | 0.138 |