Model info
| Transcription factor | STAT3 | ||||||||
| Model | STAT3_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 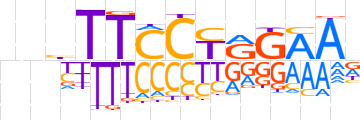 | ||||||||
| LOGO (reverse complement) | 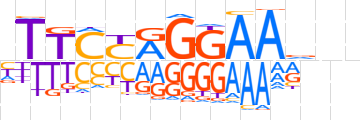 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Consensus | nnbTTCCTRGAAn | ||||||||
| wAUC | 0.8726770570749065 | ||||||||
| Best AUC | 0.9126380298922476 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 505 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT3{6.2.1.0.3} | ||||||||
| HGNC | 11364 | ||||||||
| EntrezGene | 6774 | ||||||||
| UniProt ID | STAT3_HUMAN | ||||||||
| UniProt AC | P40763 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.204 | 15.035 | 22.053 | 24.219 | 89.299 | 24.376 | 9.739 | 67.813 | 20.072 | 32.024 | 26.539 | 28.489 | 16.272 | 25.613 | 40.159 | 27.094 |
| 02 | 15.834 | 39.617 | 35.717 | 65.679 | 9.943 | 41.121 | 0.973 | 45.011 | 15.273 | 37.936 | 15.188 | 30.094 | 17.464 | 48.45 | 21.353 | 60.348 |
| 03 | 0.0 | 1.117 | 0.0 | 57.396 | 2.941 | 7.897 | 2.092 | 154.193 | 1.915 | 4.157 | 0.916 | 66.242 | 0.0 | 1.077 | 0.0 | 200.056 |
| 04 | 0.0 | 0.0 | 0.918 | 3.938 | 0.0 | 0.0 | 0.0 | 14.248 | 0.0 | 0.0 | 1.121 | 1.887 | 1.026 | 0.0 | 36.426 | 440.436 |
| 05 | 1.026 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.861 | 33.782 | 0.0 | 1.822 | 56.201 | 377.216 | 0.96 | 26.131 |
| 06 | 0.0 | 58.985 | 0.0 | 1.103 | 0.0 | 365.729 | 0.0 | 45.269 | 0.0 | 0.96 | 0.0 | 0.0 | 0.885 | 24.198 | 0.0 | 2.87 |
| 07 | 0.0 | 0.885 | 0.0 | 0.0 | 0.0 | 171.272 | 0.0 | 278.601 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 32.888 | 0.0 | 16.355 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 158.197 | 5.73 | 35.139 | 5.978 | 0.0 | 0.0 | 0.0 | 0.0 | 20.045 | 1.191 | 269.289 | 4.431 |
| 09 | 5.153 | 4.61 | 154.705 | 13.773 | 1.859 | 0.0 | 1.963 | 3.1 | 13.388 | 2.793 | 242.677 | 45.571 | 1.068 | 0.0 | 9.342 | 0.0 |
| 10 | 16.377 | 5.09 | 0.0 | 0.0 | 5.509 | 1.893 | 0.0 | 0.0 | 315.825 | 85.1 | 1.996 | 5.765 | 45.805 | 11.588 | 0.936 | 4.114 |
| 11 | 352.061 | 8.079 | 11.976 | 11.401 | 93.697 | 4.869 | 0.0 | 5.105 | 2.932 | 0.0 | 0.0 | 0.0 | 8.956 | 0.0 | 0.923 | 0.0 |
| 12 | 150.695 | 64.537 | 157.248 | 85.165 | 5.083 | 1.009 | 0.0 | 6.857 | 2.064 | 3.999 | 2.958 | 3.879 | 4.968 | 3.948 | 6.432 | 1.158 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.001 | -0.718 | -0.343 | -0.251 | 1.042 | -0.245 | -1.139 | 0.768 | -0.436 | 0.024 | -0.161 | -0.091 | -0.641 | -0.196 | 0.248 | -0.141 |
| 02 | -0.668 | 0.235 | 0.132 | 0.736 | -1.119 | 0.272 | -3.146 | 0.361 | -0.703 | 0.192 | -0.709 | -0.037 | -0.572 | 0.434 | -0.375 | 0.652 |
| 03 | -4.4 | -3.046 | -4.4 | 0.602 | -2.252 | -1.34 | -2.546 | 1.586 | -2.62 | -1.94 | -3.189 | 0.745 | -4.4 | -3.072 | -4.4 | 1.846 |
| 04 | -4.4 | -4.4 | -3.187 | -1.99 | -4.4 | -4.4 | -4.4 | -0.771 | -4.4 | -4.4 | -3.043 | -2.632 | -3.107 | -4.4 | 0.152 | 2.634 |
| 05 | -3.107 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.276 | 0.077 | -4.4 | -2.661 | 0.581 | 2.479 | -3.155 | -0.176 |
| 06 | -4.4 | 0.629 | -4.4 | -3.054 | -4.4 | 2.449 | -4.4 | 0.367 | -4.4 | -3.155 | -4.4 | -4.4 | -3.212 | -0.252 | -4.4 | -2.273 |
| 07 | -4.4 | -3.212 | -4.4 | -4.4 | -4.4 | 1.691 | -4.4 | 2.177 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 0.05 | -4.4 | -0.636 |
| 08 | -4.4 | -4.4 | -4.4 | -4.4 | 1.612 | -1.643 | 0.116 | -1.603 | -4.4 | -4.4 | -4.4 | -4.4 | -0.437 | -2.997 | 2.143 | -1.882 |
| 09 | -1.742 | -1.845 | 1.59 | -0.804 | -2.645 | -4.4 | -2.599 | -2.205 | -0.831 | -2.297 | 2.039 | 0.373 | -3.079 | -4.4 | -1.179 | -4.4 |
| 10 | -0.635 | -1.754 | -4.4 | -4.4 | -1.68 | -2.629 | -4.4 | -4.4 | 2.302 | 0.994 | -2.586 | -1.637 | 0.378 | -0.971 | -3.173 | -1.95 |
| 11 | 2.411 | -1.318 | -0.94 | -0.987 | 1.09 | -1.795 | -4.4 | -1.751 | -2.254 | -4.4 | -4.4 | -4.4 | -1.22 | -4.4 | -3.183 | -4.4 |
| 12 | 1.563 | 0.719 | 1.606 | 0.995 | -1.755 | -3.12 | -4.4 | -1.474 | -2.557 | -1.976 | -2.246 | -2.003 | -1.776 | -1.987 | -1.534 | -3.018 |