Model info
| Transcription factor | RUNX1 | ||||||||
| Model | RUNX1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 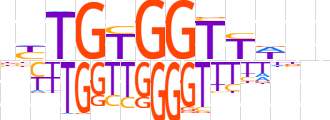 | ||||||||
| LOGO (reverse complement) | 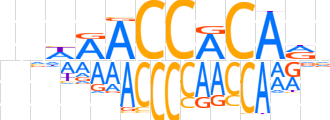 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Consensus | nYTGTGGTYWnn | ||||||||
| wAUC | 0.9293809133160168 | ||||||||
| Best AUC | 0.933601646339997 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 531 | ||||||||
| TF family | Runt-related factors{6.4.1} | ||||||||
| TF subfamily | Runx1 (PEBP2alphaB, CBF-alpha2, AML-1){6.4.1.0.2} | ||||||||
| HGNC | 10471 | ||||||||
| EntrezGene | 861 | ||||||||
| UniProt ID | RUNX1_HUMAN | ||||||||
| UniProt AC | Q01196 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.995 | 24.826 | 3.022 | 30.097 | 7.463 | 71.475 | 2.912 | 57.739 | 23.957 | 75.575 | 1.971 | 39.266 | 2.895 | 63.933 | 7.564 | 77.312 |
| 02 | 4.737 | 0.0 | 1.088 | 38.484 | 2.996 | 4.062 | 0.987 | 227.762 | 0.0 | 0.0 | 0.0 | 15.469 | 2.957 | 2.843 | 5.356 | 193.258 |
| 03 | 0.0 | 0.0 | 10.691 | 0.0 | 0.0 | 0.0 | 5.987 | 0.918 | 0.0 | 1.088 | 6.344 | 0.0 | 0.0 | 0.947 | 472.182 | 1.843 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.035 | 4.858 | 120.595 | 1.835 | 367.915 | 0.0 | 0.0 | 0.0 | 2.762 |
| 05 | 0.0 | 0.0 | 4.858 | 0.0 | 0.0 | 0.0 | 120.595 | 0.0 | 0.0 | 0.0 | 1.835 | 0.0 | 0.0 | 0.0 | 372.712 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 499.092 | 0.908 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.961 | 36.602 | 15.841 | 445.688 | 0.0 | 0.0 | 0.908 | 0.0 |
| 08 | 0.0 | 0.0 | 0.961 | 0.0 | 0.0 | 5.745 | 0.0 | 30.856 | 0.0 | 4.84 | 0.964 | 10.946 | 10.326 | 105.118 | 12.06 | 318.183 |
| 09 | 6.537 | 0.0 | 0.0 | 3.789 | 77.833 | 1.889 | 8.164 | 27.817 | 8.016 | 0.0 | 5.969 | 0.0 | 78.386 | 18.234 | 35.469 | 227.896 |
| 10 | 41.589 | 35.32 | 75.264 | 18.6 | 2.842 | 10.411 | 3.962 | 2.907 | 6.617 | 14.972 | 16.137 | 11.876 | 22.312 | 66.736 | 76.372 | 94.082 |
| 11 | 19.188 | 10.506 | 34.047 | 9.618 | 31.241 | 40.725 | 4.858 | 50.615 | 34.82 | 49.265 | 54.149 | 33.502 | 14.725 | 32.123 | 44.605 | 36.013 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.114 | -0.227 | -2.227 | -0.037 | -1.394 | 0.82 | -2.26 | 0.608 | -0.262 | 0.876 | -2.596 | 0.226 | -2.265 | 0.71 | -1.381 | 0.898 |
| 02 | -1.82 | -4.4 | -3.064 | 0.206 | -2.235 | -1.961 | -3.135 | 1.976 | -4.4 | -4.4 | -4.4 | -0.691 | -2.247 | -2.282 | -1.706 | 1.812 |
| 03 | -4.4 | -4.4 | -1.049 | -4.4 | -4.4 | -4.4 | -1.602 | -3.187 | -4.4 | -3.064 | -1.548 | -4.4 | -4.4 | -3.165 | 2.704 | -2.652 |
| 04 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.569 | -1.797 | 1.341 | -2.655 | 2.455 | -4.4 | -4.4 | -4.4 | -2.307 |
| 05 | -4.4 | -4.4 | -1.797 | -4.4 | -4.4 | -4.4 | 1.341 | -4.4 | -4.4 | -4.4 | -2.655 | -4.4 | -4.4 | -4.4 | 2.467 | -4.4 |
| 06 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.759 | -3.195 | -4.4 | -4.4 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.155 | 0.156 | -0.668 | 2.646 | -4.4 | -4.4 | -3.195 | -4.4 |
| 08 | -4.4 | -4.4 | -3.155 | -4.4 | -4.4 | -1.641 | -4.4 | -0.013 | -4.4 | -1.8 | -3.153 | -1.027 | -1.083 | 1.204 | -0.933 | 2.309 |
| 09 | -1.519 | -4.4 | -4.4 | -2.025 | 0.905 | -2.631 | -1.308 | -0.115 | -1.326 | -4.4 | -1.605 | -4.4 | 0.912 | -0.53 | 0.125 | 1.976 |
| 10 | 0.283 | 0.121 | 0.872 | -0.511 | -2.282 | -1.075 | -1.984 | -2.262 | -1.508 | -0.723 | -0.649 | -0.948 | -0.332 | 0.752 | 0.886 | 1.094 |
| 11 | -0.48 | -1.066 | 0.085 | -1.151 | -0.0 | 0.262 | -1.797 | 0.478 | 0.107 | 0.451 | 0.545 | 0.069 | -0.739 | 0.027 | 0.352 | 0.14 |