Model info
| Transcription factor | RFX5 | ||||||||
| Model | RFX5_HUMAN.H10MO.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 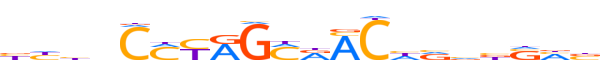 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Consensus | vdSvbbdGTYKCYRKGvvdv | ||||||||
| wAUC | 0.8269914363759591 | ||||||||
| Best AUC | 0.85237402890121 | ||||||||
| Benchmark datasets | 5 | ||||||||
| Aligned words | 494 | ||||||||
| TF family | RFX-related factors{3.3.3} | ||||||||
| TF subfamily | RFX5{3.3.3.0.5} | ||||||||
| HGNC | 9986 | ||||||||
| EntrezGene | 5993 | ||||||||
| UniProt ID | RFX5_HUMAN | ||||||||
| UniProt AC | P48382 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 84.898 | 145.734 | 231.808 | 32.23 |
| 02 | 53.744 | 50.904 | 116.102 | 273.921 |
| 03 | 45.651 | 310.206 | 77.601 | 61.213 |
| 04 | 227.988 | 130.336 | 102.839 | 33.507 |
| 05 | 41.7 | 153.244 | 147.237 | 152.489 |
| 06 | 24.914 | 263.391 | 93.018 | 113.347 |
| 07 | 182.83 | 24.217 | 85.123 | 202.5 |
| 08 | 21.371 | 4.976 | 450.334 | 17.989 |
| 09 | 4.85 | 41.279 | 57.399 | 391.142 |
| 10 | 71.23 | 100.067 | 32.797 | 290.576 |
| 11 | 78.539 | 8.373 | 326.781 | 80.977 |
| 12 | 11.428 | 440.929 | 15.19 | 27.123 |
| 13 | 7.166 | 179.391 | 7.998 | 300.116 |
| 14 | 252.971 | 10.097 | 219.957 | 11.646 |
| 15 | 63.297 | 21.192 | 335.413 | 74.769 |
| 16 | 15.855 | 18.626 | 432.51 | 27.679 |
| 17 | 100.584 | 163.712 | 165.002 | 65.372 |
| 18 | 231.493 | 94.062 | 99.562 | 69.553 |
| 19 | 141.149 | 30.583 | 248.907 | 74.031 |
| 20 | 251.141 | 79.713 | 117.333 | 46.484 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.371 | 0.162 | 0.623 | -1.31 |
| 02 | -0.817 | -0.87 | -0.062 | 0.788 |
| 03 | -0.976 | 0.912 | -0.459 | -0.691 |
| 04 | 0.606 | 0.052 | -0.182 | -1.273 |
| 05 | -1.063 | 0.212 | 0.172 | 0.207 |
| 06 | -1.554 | 0.749 | -0.281 | -0.086 |
| 07 | 0.387 | -1.581 | -0.368 | 0.488 |
| 08 | -1.698 | -2.954 | 1.283 | -1.858 |
| 09 | -2.974 | -1.073 | -0.753 | 1.143 |
| 10 | -0.543 | -0.209 | -1.294 | 0.847 |
| 11 | -0.447 | -2.535 | 0.964 | -0.417 |
| 12 | -2.267 | 1.262 | -2.012 | -1.474 |
| 13 | -2.665 | 0.368 | -2.574 | 0.879 |
| 14 | 0.709 | -2.375 | 0.57 | -2.25 |
| 15 | -0.658 | -1.706 | 0.99 | -0.495 |
| 16 | -1.973 | -1.826 | 1.243 | -1.455 |
| 17 | -0.204 | 0.277 | 0.285 | -0.627 |
| 18 | 0.621 | -0.27 | -0.214 | -0.566 |
| 19 | 0.131 | -1.36 | 0.693 | -0.505 |
| 20 | 0.702 | -0.432 | -0.052 | -0.958 |