Model info
| Transcription factor | REL | ||||||||
| Model | REL_HUMAN.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 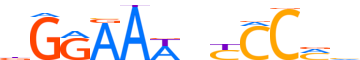 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Consensus | dKGGRnWTTCCv | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 75 | ||||||||
| TF family | NF-kappaB-related factors{6.1.1} | ||||||||
| TF subfamily | NF-kappaB p65 subunit-like factors{6.1.1.2} | ||||||||
| HGNC | 9954 | ||||||||
| EntrezGene | 5966 | ||||||||
| UniProt ID | REL_HUMAN | ||||||||
| UniProt AC | Q04864 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 18.724 | 7.125 | 23.121 | 25.377 |
| 02 | 2.113 | 10.661 | 39.726 | 21.847 |
| 03 | 0.0 | 1.85 | 71.67 | 0.827 |
| 04 | 0.735 | 2.388 | 64.033 | 7.191 |
| 05 | 25.45 | 4.146 | 41.339 | 3.412 |
| 06 | 23.222 | 21.778 | 16.32 | 13.026 |
| 07 | 23.061 | 3.595 | 1.562 | 46.128 |
| 08 | 2.113 | 0.0 | 0.0 | 72.234 |
| 09 | 0.0 | 6.994 | 2.113 | 65.24 |
| 10 | 2.205 | 55.097 | 0.735 | 16.31 |
| 11 | 0.0 | 67.536 | 0.0 | 6.81 |
| 12 | 23.796 | 28.468 | 13.758 | 8.325 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.007 | -0.874 | 0.207 | 0.297 |
| 02 | -1.819 | -0.516 | 0.73 | 0.153 |
| 03 | -2.904 | -1.905 | 1.308 | -2.335 |
| 04 | -2.384 | -1.736 | 1.197 | -0.866 |
| 05 | 0.299 | -1.326 | 0.769 | -1.477 |
| 06 | 0.212 | 0.15 | -0.122 | -0.332 |
| 07 | 0.205 | -1.437 | -2.008 | 0.876 |
| 08 | -1.819 | -2.904 | -2.904 | 1.316 |
| 09 | -2.904 | -0.89 | -1.819 | 1.216 |
| 10 | -1.79 | 1.05 | -2.384 | -0.123 |
| 11 | -2.904 | 1.25 | -2.904 | -0.914 |
| 12 | 0.235 | 0.407 | -0.282 | -0.738 |