Model info
| Transcription factor | PGR | ||||||||
| Model | PRGR_HUMAN.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 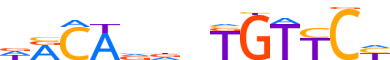 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 13 | ||||||||
| Quality | C | ||||||||
| Consensus | RGAACAnhbTGYh | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 26 | ||||||||
| TF family | Steroid hormone receptors (NR3){2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C){2.1.1.1} | ||||||||
| HGNC | 8910 | ||||||||
| EntrezGene | 5241 | ||||||||
| UniProt ID | PRGR_HUMAN | ||||||||
| UniProt AC | P06401 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 11.812 | 0.0 | 8.969 | 4.266 |
| 02 | 0.0 | 1.094 | 23.953 | 0.0 |
| 03 | 17.281 | 0.0 | 7.766 | 0.0 |
| 04 | 21.328 | 0.0 | 0.0 | 3.719 |
| 05 | 1.094 | 23.297 | 0.656 | 0.0 |
| 06 | 20.891 | 1.969 | 1.094 | 1.094 |
| 07 | 4.621 | 4.84 | 8.012 | 7.574 |
| 08 | 3.418 | 9.215 | 2.215 | 10.199 |
| 09 | 2.434 | 12.496 | 4.512 | 5.605 |
| 10 | 6.016 | 0.0 | 0.0 | 19.031 |
| 11 | 0.0 | 0.0 | 20.672 | 4.375 |
| 12 | 0.0 | 4.375 | 3.937 | 16.734 |
| 13 | 10.09 | 6.043 | 0.246 | 8.668 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.58 | -2.172 | 0.324 | -0.332 |
| 02 | -2.172 | -1.314 | 1.254 | -2.172 |
| 03 | 0.94 | -2.172 | 0.193 | -2.172 |
| 04 | 1.142 | -2.172 | -2.172 | -0.446 |
| 05 | -1.314 | 1.227 | -1.576 | -2.172 |
| 06 | 1.122 | -0.935 | -1.314 | -1.314 |
| 07 | -0.264 | -0.225 | 0.221 | 0.17 |
| 08 | -0.515 | 0.349 | -0.85 | 0.443 |
| 09 | -0.78 | 0.632 | -0.285 | -0.097 |
| 10 | -0.035 | -2.172 | -2.172 | 1.032 |
| 11 | -2.172 | -2.172 | 1.112 | -0.311 |
| 12 | -2.172 | -0.311 | -0.399 | 0.909 |
| 13 | 0.433 | -0.031 | -1.905 | 0.293 |