Model info
| Transcription factor | PPARG | ||||||||
| Model | PPARG_HUMAN.H10MO.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 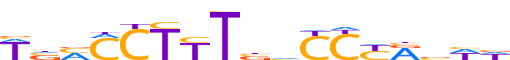 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 17 | ||||||||
| Quality | A | ||||||||
| Consensus | hWbYRGGbvARAGKKbW | ||||||||
| wAUC | 0.8598264723712874 | ||||||||
| Best AUC | 0.8693616566086644 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 499 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | PPAR (NR1C){2.1.2.5} | ||||||||
| HGNC | 9236 | ||||||||
| EntrezGene | 5468 | ||||||||
| UniProt ID | PPARG_HUMAN | ||||||||
| UniProt AC | P37231 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 286.194 | 61.228 | 57.646 | 93.667 |
| 02 | 255.195 | 15.678 | 64.472 | 163.39 |
| 03 | 52.381 | 168.558 | 180.431 | 97.364 |
| 04 | 35.532 | 77.008 | 50.313 | 335.881 |
| 05 | 193.152 | 14.487 | 255.173 | 35.922 |
| 06 | 56.065 | 0.0 | 396.699 | 45.971 |
| 07 | 78.893 | 6.725 | 388.403 | 24.714 |
| 08 | 77.863 | 77.975 | 202.256 | 140.64 |
| 09 | 67.173 | 253.421 | 117.125 | 61.015 |
| 10 | 476.554 | 7.216 | 11.922 | 3.043 |
| 11 | 345.279 | 16.988 | 116.843 | 19.625 |
| 12 | 409.316 | 2.085 | 85.457 | 1.877 |
| 13 | 67.705 | 2.078 | 415.828 | 13.125 |
| 14 | 40.353 | 2.179 | 381.853 | 74.349 |
| 15 | 17.867 | 49.231 | 186.606 | 245.031 |
| 16 | 60.92 | 276.838 | 95.515 | 65.462 |
| 17 | 370.886 | 27.728 | 46.916 | 53.204 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.824 | -0.699 | -0.757 | -0.282 |
| 02 | 0.71 | -1.991 | -0.648 | 0.267 |
| 03 | -0.85 | 0.298 | 0.366 | -0.244 |
| 04 | -1.225 | -0.474 | -0.889 | 0.983 |
| 05 | 0.433 | -2.063 | 0.71 | -1.214 |
| 06 | -0.784 | -4.398 | 1.149 | -0.977 |
| 07 | -0.451 | -2.725 | 1.128 | -1.57 |
| 08 | -0.463 | -0.462 | 0.479 | 0.119 |
| 09 | -0.608 | 0.703 | -0.062 | -0.702 |
| 10 | 1.332 | -2.667 | -2.237 | -3.313 |
| 11 | 1.011 | -1.918 | -0.064 | -1.785 |
| 12 | 1.18 | -3.547 | -0.372 | -3.606 |
| 13 | -0.6 | -3.549 | 1.196 | -2.152 |
| 14 | -1.103 | -3.521 | 1.111 | -0.509 |
| 15 | -1.872 | -0.911 | 0.399 | 0.67 |
| 16 | -0.703 | 0.791 | -0.263 | -0.633 |
| 17 | 1.082 | -1.461 | -0.957 | -0.835 |