Model info
| Transcription factor | POU5F1 | ||||||||
| Model | PO5F1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 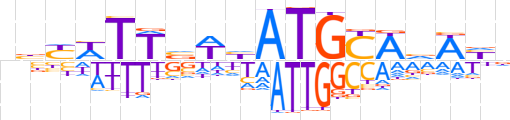 | ||||||||
| LOGO (reverse complement) | 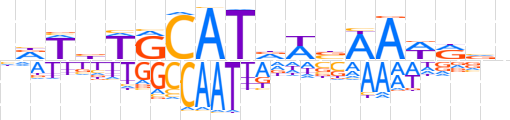 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Consensus | nbYWTTvWhATGCWvWYn | ||||||||
| wAUC | 0.9200050149283723 | ||||||||
| Best AUC | 0.9471908139850505 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 547 | ||||||||
| TF family | POU domain factors{3.1.10} | ||||||||
| TF subfamily | POU5 (Oct-3/4-like factors){3.1.10.5} | ||||||||
| HGNC | 9221 | ||||||||
| EntrezGene | 5460 | ||||||||
| UniProt ID | PO5F1_HUMAN | ||||||||
| UniProt AC | Q01860 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.598 | 31.502 | 20.518 | 13.166 | 25.92 | 59.696 | 6.974 | 36.286 | 7.793 | 68.491 | 22.203 | 25.304 | 16.233 | 70.214 | 44.094 | 39.285 |
| 02 | 2.822 | 37.14 | 0.0 | 17.581 | 18.868 | 105.546 | 1.014 | 104.475 | 4.924 | 64.177 | 7.015 | 17.673 | 5.811 | 71.451 | 15.579 | 21.2 |
| 03 | 18.655 | 1.846 | 0.972 | 10.952 | 147.575 | 14.546 | 1.283 | 114.91 | 14.885 | 4.011 | 1.768 | 2.945 | 53.43 | 7.94 | 8.079 | 91.478 |
| 04 | 5.724 | 15.783 | 5.969 | 207.07 | 0.0 | 1.828 | 0.0 | 26.515 | 0.0 | 0.862 | 0.972 | 10.267 | 4.935 | 3.796 | 0.923 | 210.631 |
| 05 | 2.847 | 0.0 | 2.843 | 4.97 | 10.82 | 0.972 | 4.642 | 5.834 | 2.952 | 0.0 | 1.896 | 3.017 | 33.963 | 1.853 | 38.265 | 380.402 |
| 06 | 1.897 | 22.024 | 16.509 | 10.152 | 0.98 | 1.846 | 0.0 | 0.0 | 6.607 | 27.795 | 7.871 | 5.371 | 49.87 | 112.384 | 204.886 | 27.082 |
| 07 | 16.231 | 0.0 | 0.0 | 43.123 | 116.447 | 1.014 | 0.0 | 46.588 | 53.826 | 2.962 | 6.916 | 165.562 | 23.794 | 0.0 | 5.936 | 12.876 |
| 08 | 46.594 | 28.533 | 12.227 | 122.944 | 0.0 | 0.884 | 0.0 | 3.092 | 1.98 | 4.094 | 2.874 | 3.905 | 36.118 | 77.136 | 27.404 | 127.491 |
| 09 | 80.777 | 0.0 | 1.789 | 2.125 | 105.881 | 0.0 | 0.0 | 4.766 | 40.651 | 0.0 | 0.949 | 0.904 | 246.9 | 0.949 | 2.951 | 6.632 |
| 10 | 5.77 | 0.0 | 11.736 | 456.704 | 0.0 | 0.0 | 0.0 | 0.949 | 0.0 | 0.0 | 0.0 | 5.689 | 0.0 | 0.0 | 0.0 | 14.428 |
| 11 | 0.941 | 0.0 | 3.728 | 1.102 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.862 | 0.874 | 5.242 | 0.0 | 443.141 | 29.386 |
| 12 | 0.0 | 6.182 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.98 | 349.367 | 36.971 | 70.413 | 0.0 | 24.937 | 0.0 | 6.424 |
| 13 | 0.98 | 0.0 | 0.0 | 0.0 | 300.152 | 5.846 | 40.03 | 34.459 | 25.033 | 0.0 | 2.913 | 9.025 | 62.391 | 0.0 | 0.914 | 13.532 |
| 14 | 202.994 | 50.526 | 75.57 | 59.466 | 1.988 | 2.917 | 0.0 | 0.941 | 2.951 | 33.297 | 4.775 | 2.834 | 37.397 | 0.0 | 9.853 | 9.766 |
| 15 | 204.417 | 7.959 | 17.396 | 15.557 | 71.115 | 4.495 | 3.151 | 7.98 | 69.222 | 5.772 | 7.212 | 7.991 | 39.031 | 0.972 | 16.541 | 16.462 |
| 16 | 56.041 | 41.824 | 32.035 | 253.885 | 2.942 | 7.744 | 1.179 | 7.335 | 7.962 | 14.78 | 9.258 | 12.3 | 3.842 | 10.413 | 8.706 | 25.03 |
| 17 | 25.168 | 10.899 | 12.249 | 22.471 | 37.267 | 14.844 | 6.895 | 15.755 | 10.043 | 19.595 | 11.637 | 9.903 | 70.065 | 39.449 | 102.605 | 86.431 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.367 | 0.017 | -0.405 | -0.838 | -0.175 | 0.651 | -1.449 | 0.157 | -1.343 | 0.787 | -0.327 | -0.199 | -0.634 | 0.812 | 0.35 | 0.236 |
| 02 | -2.279 | 0.18 | -4.392 | -0.556 | -0.487 | 1.218 | -3.107 | 1.208 | -1.775 | 0.723 | -1.443 | -0.551 | -1.621 | 0.829 | -0.674 | -0.373 |
| 03 | -0.498 | -2.641 | -3.137 | -1.017 | 1.552 | -0.741 | -2.932 | 1.303 | -0.719 | -1.964 | -2.677 | -2.241 | 0.541 | -1.325 | -1.309 | 1.075 |
| 04 | -1.635 | -0.662 | -1.595 | 1.89 | -4.392 | -2.65 | -4.392 | -0.153 | -4.392 | -3.222 | -3.137 | -1.079 | -1.773 | -2.014 | -3.174 | 1.907 |
| 05 | -2.271 | -4.392 | -2.272 | -1.766 | -1.028 | -3.137 | -1.83 | -1.617 | -2.239 | -4.392 | -2.619 | -2.22 | 0.092 | -2.638 | 0.21 | 2.497 |
| 06 | -2.619 | -0.335 | -0.618 | -1.09 | -3.132 | -2.641 | -4.392 | -4.392 | -1.5 | -0.106 | -1.334 | -1.694 | 0.472 | 1.28 | 1.879 | -0.132 |
| 07 | -0.634 | -4.392 | -4.392 | 0.328 | 1.316 | -3.107 | -4.392 | 0.405 | 0.548 | -2.236 | -1.457 | 1.667 | -0.259 | -4.392 | -1.601 | -0.86 |
| 08 | 0.405 | -0.08 | -0.91 | 1.37 | -4.392 | -3.204 | -4.392 | -2.198 | -2.583 | -1.945 | -2.263 | -1.988 | 0.152 | 0.906 | -0.12 | 1.406 |
| 09 | 0.952 | -4.392 | -2.667 | -2.523 | 1.221 | -4.392 | -4.392 | -1.805 | 0.27 | -4.392 | -3.155 | -3.189 | 2.066 | -3.155 | -2.239 | -1.496 |
| 10 | -1.627 | -4.392 | -0.95 | 2.68 | -4.392 | -4.392 | -4.392 | -3.155 | -4.392 | -4.392 | -4.392 | -1.641 | -4.392 | -4.392 | -4.392 | -0.749 |
| 11 | -3.161 | -4.392 | -2.03 | -3.046 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -1.025 | -3.213 | -1.717 | -4.392 | 2.65 | -0.051 |
| 12 | -4.392 | -1.562 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -4.392 | -3.132 | 2.412 | 0.176 | 0.815 | -4.392 | -0.213 | -4.392 | -1.526 |
| 13 | -3.132 | -4.392 | -4.392 | -4.392 | 2.261 | -1.615 | 0.254 | 0.106 | -0.209 | -4.392 | -2.251 | -1.203 | 0.695 | -4.392 | -3.181 | -0.812 |
| 14 | 1.87 | 0.485 | 0.885 | 0.647 | -2.58 | -2.249 | -4.392 | -3.161 | -2.239 | 0.072 | -1.804 | -2.275 | 0.187 | -4.392 | -1.119 | -1.127 |
| 15 | 1.877 | -1.323 | -0.567 | -0.676 | 0.825 | -1.859 | -2.181 | -1.321 | 0.798 | -1.627 | -1.417 | -1.319 | 0.229 | -3.137 | -0.616 | -0.621 |
| 16 | 0.588 | 0.298 | 0.034 | 2.093 | -2.242 | -1.349 | -2.996 | -1.401 | -1.323 | -0.726 | -1.178 | -0.904 | -2.003 | -1.065 | -1.237 | -0.21 |
| 17 | -0.204 | -1.021 | -0.908 | -0.316 | 0.183 | -0.722 | -1.459 | -0.664 | -1.1 | -0.45 | -0.958 | -1.114 | 0.81 | 0.24 | 1.19 | 1.019 |