Model info
| Transcription factor | TP73 | ||||||||
| Model | P73_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 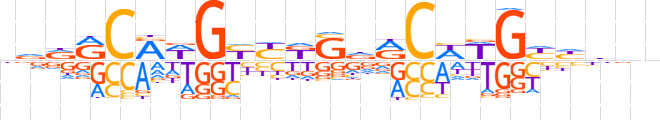 | ||||||||
| LOGO (reverse complement) | 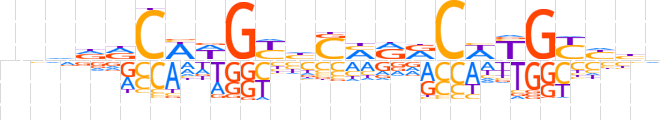 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nvdRCRWGYYKSdRCWWGYhbnn | ||||||||
| wAUC | 0.9876071808481912 | ||||||||
| Best AUC | 0.9944282470243889 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 490 | ||||||||
| TF family | p53-related factors{6.3.1} | ||||||||
| TF subfamily | p73{6.3.1.0.3} | ||||||||
| HGNC | 12003 | ||||||||
| EntrezGene | 7161 | ||||||||
| UniProt ID | P73_HUMAN | ||||||||
| UniProt AC | O15350 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 25.76 | 9.056 | 54.214 | 10.735 | 72.571 | 20.081 | 24.724 | 25.888 | 24.844 | 15.624 | 52.526 | 11.095 | 4.793 | 20.178 | 93.874 | 16.097 |
| 02 | 18.394 | 8.89 | 90.47 | 10.215 | 21.825 | 3.039 | 4.134 | 35.941 | 47.88 | 10.598 | 144.279 | 22.582 | 7.863 | 6.365 | 35.849 | 13.737 |
| 03 | 53.728 | 1.094 | 38.377 | 2.762 | 20.805 | 0.0 | 3.106 | 4.981 | 78.159 | 0.962 | 191.364 | 4.246 | 18.934 | 1.014 | 53.889 | 8.638 |
| 04 | 1.879 | 160.699 | 4.967 | 4.081 | 0.0 | 3.069 | 0.0 | 0.0 | 1.909 | 281.056 | 0.962 | 2.809 | 0.0 | 18.52 | 0.0 | 2.107 |
| 05 | 1.909 | 0.933 | 0.946 | 0.0 | 354.689 | 33.357 | 43.601 | 31.698 | 1.103 | 4.826 | 0.0 | 0.0 | 2.009 | 1.852 | 3.087 | 2.05 |
| 06 | 114.225 | 7.011 | 60.873 | 177.601 | 5.701 | 1.109 | 0.0 | 34.158 | 8.03 | 0.925 | 0.0 | 38.678 | 6.872 | 0.0 | 0.0 | 26.877 |
| 07 | 0.0 | 0.0 | 134.828 | 0.0 | 0.0 | 0.0 | 9.046 | 0.0 | 0.939 | 0.0 | 58.988 | 0.946 | 0.0 | 0.0 | 277.313 | 0.0 |
| 08 | 0.939 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 31.978 | 180.87 | 19.445 | 247.882 | 0.0 | 0.0 | 0.0 | 0.946 |
| 09 | 7.17 | 12.82 | 0.0 | 12.928 | 14.001 | 126.528 | 3.066 | 37.276 | 0.0 | 5.691 | 0.0 | 13.754 | 16.061 | 124.26 | 11.434 | 97.073 |
| 10 | 11.1 | 0.0 | 11.808 | 14.323 | 58.029 | 3.977 | 6.215 | 201.078 | 4.722 | 0.0 | 2.943 | 6.835 | 15.386 | 2.134 | 96.355 | 47.155 |
| 11 | 6.116 | 18.001 | 58.316 | 6.804 | 0.0 | 0.0 | 6.111 | 0.0 | 4.26 | 11.004 | 101.12 | 0.936 | 20.491 | 36.503 | 203.697 | 8.701 |
| 12 | 0.96 | 3.904 | 26.003 | 0.0 | 30.39 | 3.877 | 3.143 | 28.098 | 82.362 | 39.887 | 200.999 | 45.995 | 1.069 | 0.949 | 11.564 | 2.86 |
| 13 | 52.983 | 0.885 | 58.892 | 2.02 | 32.039 | 0.0 | 3.754 | 12.823 | 98.378 | 0.0 | 127.251 | 16.079 | 30.539 | 0.0 | 37.215 | 9.2 |
| 14 | 1.945 | 209.966 | 0.0 | 2.028 | 0.0 | 0.885 | 0.0 | 0.0 | 0.0 | 226.065 | 0.0 | 1.048 | 0.0 | 40.122 | 0.0 | 0.0 |
| 15 | 1.945 | 0.0 | 0.0 | 0.0 | 263.701 | 53.731 | 6.16 | 153.447 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.076 | 0.0 | 0.0 |
| 16 | 39.656 | 22.722 | 42.485 | 160.783 | 2.102 | 1.006 | 0.0 | 53.699 | 0.0 | 0.0 | 2.155 | 4.006 | 14.432 | 5.985 | 5.926 | 127.103 |
| 17 | 1.054 | 2.094 | 53.042 | 0.0 | 6.131 | 0.0 | 23.581 | 0.0 | 7.666 | 0.0 | 41.03 | 1.871 | 1.917 | 0.0 | 343.675 | 0.0 |
| 18 | 2.007 | 2.904 | 3.977 | 7.88 | 0.898 | 1.197 | 0.0 | 0.0 | 53.87 | 218.582 | 12.858 | 176.018 | 0.0 | 0.0 | 0.0 | 1.871 |
| 19 | 13.28 | 20.976 | 8.458 | 14.06 | 37.778 | 140.929 | 4.828 | 39.147 | 1.18 | 8.587 | 3.124 | 3.945 | 21.656 | 82.6 | 15.343 | 66.17 |
| 20 | 14.195 | 19.228 | 28.511 | 11.959 | 37.657 | 73.198 | 9.108 | 133.129 | 3.116 | 3.867 | 5.986 | 18.785 | 10.615 | 33.768 | 46.412 | 32.527 |
| 21 | 11.193 | 14.595 | 29.194 | 10.602 | 51.599 | 30.593 | 12.837 | 35.032 | 20.046 | 25.092 | 30.391 | 14.487 | 19.636 | 28.138 | 112.635 | 35.991 |
| 22 | 23.718 | 21.935 | 40.329 | 16.492 | 31.602 | 29.26 | 2.8 | 34.756 | 25.963 | 39.991 | 58.204 | 60.899 | 11.41 | 20.61 | 31.255 | 32.836 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.155 | -1.173 | 0.582 | -1.009 | 0.872 | -0.399 | -0.195 | -0.15 | -0.19 | -0.645 | 0.55 | -0.978 | -1.774 | -0.395 | 1.128 | -0.616 |
| 02 | -0.485 | -1.191 | 1.091 | -1.057 | -0.318 | -2.187 | -1.91 | 0.174 | 0.459 | -1.022 | 1.556 | -0.284 | -1.308 | -1.509 | 0.172 | -0.77 |
| 03 | 0.573 | -3.026 | 0.239 | -2.271 | -0.365 | -4.37 | -2.168 | -1.738 | 0.945 | -3.12 | 1.838 | -1.885 | -0.457 | -3.082 | 0.576 | -1.218 |
| 04 | -2.601 | 1.664 | -1.741 | -1.921 | -4.37 | -2.178 | -4.37 | -4.37 | -2.587 | 2.222 | -3.12 | -2.256 | -4.37 | -0.479 | -4.37 | -2.504 |
| 05 | -2.587 | -3.141 | -3.132 | -4.37 | 2.454 | 0.101 | 0.366 | 0.05 | -3.02 | -1.767 | -4.37 | -4.37 | -2.545 | -2.613 | -2.173 | -2.528 |
| 06 | 1.323 | -1.417 | 0.697 | 1.763 | -1.612 | -3.016 | -4.37 | 0.124 | -1.288 | -3.147 | -4.37 | 0.247 | -1.436 | -4.37 | -4.37 | -0.113 |
| 07 | -4.37 | -4.37 | 1.489 | -4.37 | -4.37 | -4.37 | -1.174 | -4.37 | -3.136 | -4.37 | 0.666 | -3.132 | -4.37 | -4.37 | 2.208 | -4.37 |
| 08 | -3.136 | -4.37 | -4.37 | -4.37 | -4.37 | -4.37 | -4.37 | -4.37 | 0.059 | 1.782 | -0.431 | 2.096 | -4.37 | -4.37 | -4.37 | -3.132 |
| 09 | -1.396 | -0.838 | -4.37 | -0.829 | -0.752 | 1.425 | -2.179 | 0.21 | -4.37 | -1.614 | -4.37 | -0.769 | -0.618 | 1.407 | -0.948 | 1.161 |
| 10 | -0.977 | -4.37 | -0.917 | -0.73 | 0.649 | -1.945 | -1.531 | 1.887 | -1.787 | -4.37 | -2.215 | -1.441 | -0.66 | -2.494 | 1.154 | 0.443 |
| 11 | -1.546 | -0.507 | 0.654 | -1.446 | -4.37 | -4.37 | -1.547 | -4.37 | -1.882 | -0.985 | 1.202 | -3.139 | -0.38 | 0.19 | 1.9 | -1.211 |
| 12 | -3.121 | -1.962 | -0.145 | -4.37 | 0.009 | -1.968 | -2.157 | -0.069 | 0.998 | 0.277 | 1.887 | 0.419 | -3.043 | -3.129 | -0.938 | -2.241 |
| 13 | 0.559 | -3.178 | 0.664 | -2.54 | 0.061 | -4.37 | -1.998 | -0.837 | 1.175 | -4.37 | 1.431 | -0.617 | 0.013 | -4.37 | 0.209 | -1.158 |
| 14 | -2.572 | 1.931 | -4.37 | -2.537 | -4.37 | -3.178 | -4.37 | -4.37 | -4.37 | 2.004 | -4.37 | -3.058 | -4.37 | 0.283 | -4.37 | -4.37 |
| 15 | -2.572 | -4.37 | -4.37 | -4.37 | 2.158 | 0.573 | -1.539 | 1.618 | -4.37 | -4.37 | -4.37 | -4.37 | -4.37 | -2.176 | -4.37 | -4.37 |
| 16 | 0.272 | -0.278 | 0.34 | 1.664 | -2.507 | -3.088 | -4.37 | 0.572 | -4.37 | -4.37 | -2.486 | -1.938 | -0.722 | -1.567 | -1.576 | 1.43 |
| 17 | -3.053 | -2.51 | 0.56 | -4.37 | -1.544 | -4.37 | -0.242 | -4.37 | -1.332 | -4.37 | 0.305 | -2.604 | -2.584 | -4.37 | 2.423 | -4.37 |
| 18 | -2.546 | -2.227 | -1.945 | -1.306 | -3.168 | -2.959 | -4.37 | -4.37 | 0.575 | 1.971 | -0.835 | 1.755 | -4.37 | -4.37 | -4.37 | -2.604 |
| 19 | -0.803 | -0.357 | -1.238 | -0.748 | 0.224 | 1.533 | -1.767 | 0.259 | -2.97 | -1.224 | -2.163 | -1.952 | -0.325 | 1.0 | -0.663 | 0.78 |
| 20 | -0.739 | -0.442 | -0.054 | -0.905 | 0.221 | 0.88 | -1.168 | 1.476 | -2.165 | -1.971 | -1.566 | -0.465 | -1.02 | 0.113 | 0.428 | 0.076 |
| 21 | -0.969 | -0.711 | -0.031 | -1.021 | 0.533 | 0.015 | -0.836 | 0.149 | -0.401 | -0.18 | 0.009 | -0.719 | -0.421 | -0.067 | 1.309 | 0.176 |
| 22 | -0.236 | -0.313 | 0.288 | -0.592 | 0.047 | -0.029 | -2.259 | 0.141 | -0.147 | 0.28 | 0.652 | 0.697 | -0.95 | -0.374 | 0.036 | 0.085 |