Model info
| Transcription factor | TP53 | ||||||||
| Model | P53_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 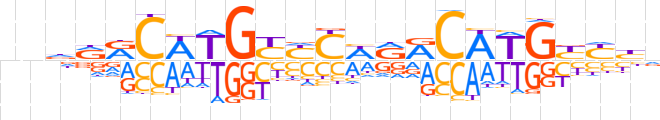 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | B | ||||||||
| Consensus | nvRRCWTGYYWGRRCAWGYYhnn | ||||||||
| wAUC | 0.8325890243818966 | ||||||||
| Best AUC | 0.9466958564195238 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 522 | ||||||||
| TF family | p53-related factors{6.3.1} | ||||||||
| TF subfamily | p53{6.3.1.0.1} | ||||||||
| HGNC | 11998 | ||||||||
| EntrezGene | 7157 | ||||||||
| UniProt ID | P53_HUMAN | ||||||||
| UniProt AC | P04637 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 44.152 | 6.615 | 55.751 | 7.657 | 97.673 | 13.058 | 18.554 | 9.363 | 33.211 | 8.542 | 52.534 | 9.195 | 16.471 | 10.635 | 101.346 | 11.762 |
| 02 | 40.053 | 7.3 | 137.65 | 6.502 | 16.684 | 2.645 | 4.817 | 14.704 | 46.139 | 8.57 | 144.836 | 28.64 | 7.688 | 0.0 | 22.735 | 7.555 |
| 03 | 78.081 | 8.696 | 22.822 | 0.965 | 11.039 | 5.682 | 0.0 | 1.794 | 102.404 | 18.381 | 175.306 | 13.947 | 15.448 | 0.858 | 37.187 | 3.908 |
| 04 | 6.665 | 187.921 | 2.684 | 9.703 | 6.952 | 13.855 | 4.002 | 8.808 | 0.917 | 227.242 | 4.202 | 2.954 | 0.927 | 15.083 | 1.789 | 2.815 |
| 05 | 6.662 | 0.887 | 1.239 | 6.671 | 368.773 | 5.424 | 14.576 | 55.328 | 2.644 | 1.031 | 4.41 | 4.592 | 9.003 | 0.862 | 8.844 | 5.572 |
| 06 | 45.664 | 12.775 | 13.355 | 315.288 | 1.739 | 0.0 | 0.0 | 6.464 | 3.764 | 0.0 | 3.1 | 22.206 | 7.121 | 0.939 | 10.298 | 53.805 |
| 07 | 0.852 | 0.0 | 55.201 | 2.235 | 0.92 | 1.976 | 10.818 | 0.0 | 0.0 | 4.134 | 15.175 | 7.444 | 0.0 | 2.153 | 392.143 | 3.467 |
| 08 | 0.0 | 1.771 | 0.0 | 0.0 | 1.976 | 5.362 | 0.0 | 0.925 | 17.179 | 142.407 | 11.048 | 302.703 | 0.852 | 4.197 | 0.0 | 8.097 |
| 09 | 1.908 | 13.288 | 0.902 | 3.909 | 14.733 | 117.41 | 0.0 | 21.595 | 0.0 | 4.871 | 0.0 | 6.177 | 17.468 | 171.62 | 18.702 | 103.936 |
| 10 | 6.09 | 1.927 | 8.772 | 17.32 | 60.96 | 13.473 | 5.9 | 226.855 | 3.884 | 0.0 | 1.142 | 14.578 | 24.674 | 4.004 | 55.401 | 51.536 |
| 11 | 20.569 | 8.802 | 62.289 | 3.947 | 0.0 | 0.0 | 19.404 | 0.0 | 9.7 | 6.114 | 55.402 | 0.0 | 36.223 | 17.543 | 256.524 | 0.0 |
| 12 | 8.324 | 1.019 | 54.26 | 2.889 | 9.2 | 3.964 | 6.411 | 12.884 | 51.909 | 24.339 | 266.869 | 50.502 | 2.834 | 0.0 | 1.114 | 0.0 |
| 13 | 39.708 | 2.727 | 29.831 | 0.0 | 24.811 | 1.778 | 1.832 | 0.902 | 132.272 | 3.259 | 189.279 | 3.843 | 23.322 | 0.837 | 33.719 | 8.398 |
| 14 | 1.148 | 215.212 | 0.977 | 2.775 | 0.0 | 3.729 | 0.0 | 4.872 | 1.898 | 252.763 | 0.0 | 0.0 | 0.0 | 13.143 | 0.0 | 0.0 |
| 15 | 0.887 | 1.011 | 1.148 | 0.0 | 388.382 | 11.177 | 8.83 | 76.458 | 0.0 | 0.0 | 0.0 | 0.977 | 0.998 | 3.622 | 0.85 | 2.178 |
| 16 | 80.196 | 10.547 | 25.739 | 273.785 | 0.928 | 0.884 | 0.908 | 13.091 | 1.148 | 0.0 | 0.884 | 8.796 | 11.796 | 1.04 | 2.721 | 64.055 |
| 17 | 12.717 | 1.851 | 79.5 | 0.0 | 4.814 | 0.0 | 6.78 | 0.876 | 2.797 | 0.88 | 24.787 | 1.788 | 8.493 | 2.211 | 346.356 | 2.668 |
| 18 | 1.86 | 2.937 | 5.778 | 18.246 | 2.031 | 1.117 | 0.0 | 1.794 | 26.089 | 198.2 | 1.819 | 231.315 | 0.0 | 1.709 | 1.884 | 1.739 |
| 19 | 3.063 | 17.475 | 3.829 | 5.612 | 24.344 | 146.345 | 2.089 | 31.185 | 0.898 | 2.889 | 4.765 | 0.929 | 31.461 | 127.338 | 13.984 | 80.311 |
| 20 | 8.759 | 15.623 | 13.408 | 21.977 | 73.42 | 110.108 | 4.472 | 106.047 | 4.947 | 9.112 | 1.807 | 8.8 | 30.006 | 44.04 | 13.647 | 30.345 |
| 21 | 9.623 | 31.368 | 63.391 | 12.749 | 71.819 | 44.885 | 17.075 | 45.104 | 3.664 | 8.445 | 14.579 | 6.646 | 32.476 | 25.432 | 79.353 | 29.908 |
| 22 | 15.136 | 37.291 | 49.783 | 15.372 | 30.762 | 45.325 | 6.424 | 27.618 | 17.612 | 43.23 | 72.899 | 40.657 | 9.444 | 21.02 | 38.352 | 25.593 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.349 | -1.501 | 0.58 | -1.362 | 1.138 | -0.849 | -0.506 | -1.17 | 0.067 | -1.258 | 0.521 | -1.187 | -0.623 | -1.047 | 1.175 | -0.95 |
| 02 | 0.252 | -1.408 | 1.48 | -1.517 | -0.61 | -2.338 | -1.798 | -0.733 | 0.393 | -1.255 | 1.531 | -0.079 | -1.359 | -4.394 | -0.307 | -1.375 |
| 03 | 0.915 | -1.241 | -0.303 | -3.145 | -1.012 | -1.644 | -4.394 | -2.667 | 1.185 | -0.515 | 1.721 | -0.785 | -0.685 | -3.228 | 0.179 | -1.99 |
| 04 | -1.494 | 1.791 | -2.325 | -1.136 | -1.454 | -0.791 | -1.968 | -1.229 | -3.181 | 1.98 | -1.924 | -2.241 | -3.174 | -0.709 | -2.67 | -2.283 |
| 05 | -1.494 | -3.205 | -2.961 | -1.493 | 2.464 | -1.688 | -0.742 | 0.573 | -2.338 | -3.098 | -1.879 | -1.842 | -1.208 | -3.225 | -1.225 | -1.662 |
| 06 | 0.382 | -0.87 | -0.827 | 2.307 | -2.693 | -4.394 | -4.394 | -1.523 | -2.024 | -4.394 | -2.198 | -0.33 | -1.431 | -3.165 | -1.079 | 0.545 |
| 07 | -3.233 | -4.394 | 0.571 | -2.483 | -3.179 | -2.587 | -1.031 | -4.394 | -4.394 | -1.938 | -0.703 | -1.389 | -4.394 | -2.515 | 2.525 | -2.098 |
| 08 | -4.394 | -2.678 | -4.394 | -4.394 | -2.587 | -1.698 | -4.394 | -3.175 | -0.581 | 1.514 | -1.011 | 2.267 | -3.233 | -1.925 | -4.394 | -1.309 |
| 09 | -2.616 | -0.832 | -3.193 | -1.99 | -0.731 | 1.322 | -4.394 | -0.357 | -4.394 | -1.788 | -4.394 | -1.566 | -0.565 | 1.7 | -0.498 | 1.2 |
| 10 | -1.579 | -2.608 | -1.233 | -0.573 | 0.669 | -0.818 | -1.609 | 1.979 | -1.995 | -4.394 | -3.022 | -0.742 | -0.226 | -1.968 | 0.574 | 0.502 |
| 11 | -0.405 | -1.229 | 0.691 | -1.981 | -4.394 | -4.394 | -0.462 | -4.394 | -1.136 | -1.575 | 0.574 | -4.394 | 0.153 | -0.561 | 2.101 | -4.394 |
| 12 | -1.283 | -3.106 | 0.553 | -2.26 | -1.187 | -1.977 | -1.531 | -0.862 | 0.509 | -0.24 | 2.141 | 0.482 | -2.278 | -4.394 | -3.041 | -4.394 |
| 13 | 0.244 | -2.311 | -0.039 | -4.394 | -0.221 | -2.674 | -2.65 | -3.193 | 1.44 | -2.154 | 1.798 | -2.005 | -0.282 | -3.245 | 0.082 | -1.274 |
| 14 | -3.018 | 1.926 | -3.136 | -2.296 | -4.394 | -2.032 | -4.394 | -1.787 | -2.62 | 2.087 | -4.394 | -4.394 | -4.394 | -0.842 | -4.394 | -4.394 |
| 15 | -3.205 | -3.111 | -3.018 | -4.394 | 2.516 | -0.999 | -1.226 | 0.894 | -4.394 | -4.394 | -4.394 | -3.136 | -3.121 | -2.059 | -3.234 | -2.505 |
| 16 | 0.942 | -1.055 | -0.184 | 2.166 | -3.173 | -3.207 | -3.188 | -0.846 | -3.018 | -4.394 | -3.207 | -1.23 | -0.947 | -3.091 | -2.313 | 0.718 |
| 17 | -0.874 | -2.641 | 0.933 | -4.394 | -1.798 | -4.394 | -1.478 | -3.213 | -2.289 | -3.21 | -0.222 | -2.67 | -1.264 | -2.492 | 2.401 | -2.33 |
| 18 | -2.638 | -2.246 | -1.628 | -0.522 | -2.564 | -3.039 | -4.394 | -2.667 | -0.171 | 1.844 | -2.656 | 1.998 | -4.394 | -2.707 | -2.627 | -2.693 |
| 19 | -2.209 | -0.565 | -2.008 | -1.656 | -0.239 | 1.541 | -2.541 | 0.005 | -3.196 | -2.261 | -1.808 | -3.172 | 0.014 | 1.402 | -0.782 | 0.943 |
| 20 | -1.234 | -0.674 | -0.823 | -0.34 | 0.854 | 1.258 | -1.866 | 1.22 | -1.773 | -1.196 | -2.661 | -1.23 | -0.033 | 0.346 | -0.806 | -0.022 |
| 21 | -1.144 | 0.011 | 0.708 | -0.872 | 0.832 | 0.365 | -0.587 | 0.37 | -2.048 | -1.269 | -0.742 | -1.497 | 0.045 | -0.196 | 0.931 | -0.036 |
| 22 | -0.705 | 0.182 | 0.468 | -0.69 | -0.009 | 0.375 | -1.529 | -0.115 | -0.557 | 0.328 | 0.847 | 0.267 | -1.162 | -0.384 | 0.209 | -0.19 |