Model info
| Transcription factor | MEOX1 | ||||||||
| Model | MEOX1_HUMAN.H10MO.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 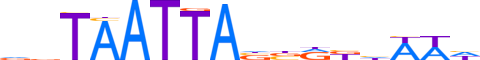 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HT-SELEX | ||||||||
| Model length | 16 | ||||||||
| Quality | D | ||||||||
| Consensus | nbTAATTAnbRbhWWh | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 61 | ||||||||
| TF family | HOX-related factors{3.1.1} | ||||||||
| TF subfamily | MEOX{3.1.1.14} | ||||||||
| HGNC | 7013 | ||||||||
| EntrezGene | 4222 | ||||||||
| UniProt ID | MEOX1_HUMAN | ||||||||
| UniProt AC | P50221 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 13.5 | 9.5 | 28.5 | 9.5 |
| 02 | 10.75 | 11.75 | 25.75 | 12.75 |
| 03 | 0.75 | 1.75 | 0.75 | 57.75 |
| 04 | 54.75 | 3.75 | 0.75 | 1.75 |
| 05 | 61.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 61.0 |
| 07 | 0.0 | 0.0 | 3.0 | 58.0 |
| 08 | 60.0 | 0.0 | 1.0 | 0.0 |
| 09 | 7.0 | 15.0 | 32.0 | 7.0 |
| 10 | 2.0 | 25.0 | 21.0 | 13.0 |
| 11 | 15.0 | 3.0 | 35.0 | 8.0 |
| 12 | 5.0 | 6.0 | 18.0 | 32.0 |
| 13 | 18.25 | 14.25 | 5.25 | 23.25 |
| 14 | 41.5 | 3.5 | 0.5 | 15.5 |
| 15 | 32.5 | 0.5 | 1.5 | 26.5 |
| 16 | 24.25 | 7.25 | 4.25 | 25.25 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.114 | -0.436 | 0.596 | -0.436 |
| 02 | -0.324 | -0.242 | 0.498 | -0.167 |
| 03 | -2.214 | -1.768 | -2.214 | 1.284 |
| 04 | 1.232 | -1.226 | -2.214 | -1.768 |
| 05 | 1.338 | -2.762 | -2.762 | -2.762 |
| 06 | -2.762 | -2.762 | -2.762 | 1.338 |
| 07 | -2.762 | -2.762 | -1.397 | 1.288 |
| 08 | 1.322 | -2.762 | -2.083 | -2.762 |
| 09 | -0.707 | -0.015 | 0.708 | -0.707 |
| 10 | -1.682 | 0.469 | 0.303 | -0.149 |
| 11 | -0.015 | -1.397 | 0.794 | -0.589 |
| 12 | -0.993 | -0.84 | 0.156 | 0.708 |
| 13 | 0.169 | -0.063 | -0.953 | 0.4 |
| 14 | 0.96 | -1.28 | -2.366 | 0.015 |
| 15 | 0.723 | -2.366 | -1.862 | 0.525 |
| 16 | 0.44 | -0.676 | -1.126 | 0.479 |