Model info
| Transcription factor | MEIS1 | ||||||||
| Model | MEIS1_HUMAN.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 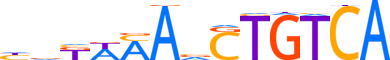 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 13 | ||||||||
| Quality | C | ||||||||
| Consensus | TGACAGbTKWMhd | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 40 | ||||||||
| TF family | TALE-type homeo domain factors{3.1.4} | ||||||||
| TF subfamily | MEIS{3.1.4.2} | ||||||||
| HGNC | 7000 | ||||||||
| EntrezGene | 4211 | ||||||||
| UniProt ID | MEIS1_HUMAN | ||||||||
| UniProt AC | O00470 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.0 | 0.0 | 0.0 | 39.347 |
| 02 | 0.968 | 0.0 | 38.379 | 0.0 |
| 03 | 36.444 | 0.0 | 0.968 | 1.935 |
| 04 | 0.968 | 36.659 | 0.0 | 1.72 |
| 05 | 36.552 | 0.968 | 0.968 | 0.86 |
| 06 | 0.0 | 5.805 | 31.606 | 1.935 |
| 07 | 2.795 | 10.643 | 9.568 | 16.341 |
| 08 | 0.0 | 0.968 | 0.0 | 38.379 |
| 09 | 0.0 | 2.903 | 8.6 | 27.844 |
| 10 | 12.363 | 2.903 | 0.968 | 23.114 |
| 11 | 24.941 | 7.74 | 3.763 | 2.903 |
| 12 | 10.535 | 12.471 | 3.763 | 12.578 |
| 13 | 9.326 | 3.628 | 20.937 | 5.456 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -2.461 | -2.461 | -2.461 | 1.32 |
| 02 | -1.741 | -2.461 | 1.296 | -2.461 |
| 03 | 1.245 | -2.461 | -1.741 | -1.327 |
| 04 | -1.741 | 1.251 | -2.461 | -1.405 |
| 05 | 1.248 | -1.741 | -1.741 | -1.8 |
| 06 | -2.461 | -0.47 | 1.107 | -1.327 |
| 07 | -1.063 | 0.072 | -0.025 | 0.473 |
| 08 | -2.461 | -1.741 | -2.461 | 1.296 |
| 09 | -2.461 | -1.035 | -0.122 | 0.984 |
| 10 | 0.211 | -1.035 | -1.741 | 0.804 |
| 11 | 0.877 | -0.217 | -0.832 | -1.035 |
| 12 | 0.063 | 0.219 | -0.832 | 0.227 |
| 13 | -0.049 | -0.861 | 0.709 | -0.523 |