Model info
| Transcription factor | MEF2A | ||||||||
| Model | MEF2A_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 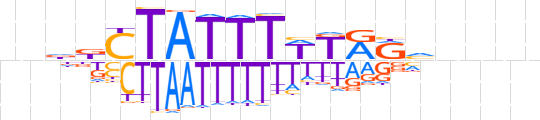 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | nnddCTATTTWKRRhnnnn | ||||||||
| wAUC | 0.8877238426104118 | ||||||||
| Best AUC | 0.9305701982712395 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 442 | ||||||||
| TF family | Regulators of differentiation{5.1.1} | ||||||||
| TF subfamily | MEF-2{5.1.1.1} | ||||||||
| HGNC | 6993 | ||||||||
| EntrezGene | 4205 | ||||||||
| UniProt ID | MEF2A_HUMAN | ||||||||
| UniProt AC | Q02078 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 35.811 | 11.861 | 41.535 | 25.416 | 27.196 | 9.909 | 5.781 | 29.458 | 31.806 | 33.932 | 33.645 | 36.446 | 21.589 | 11.33 | 61.691 | 19.493 |
| 02 | 31.088 | 9.403 | 40.161 | 35.751 | 26.814 | 4.009 | 9.316 | 26.894 | 47.649 | 10.187 | 42.03 | 42.785 | 11.651 | 5.098 | 38.695 | 55.369 |
| 03 | 15.245 | 6.987 | 58.458 | 36.512 | 4.853 | 3.65 | 6.666 | 13.528 | 8.941 | 10.584 | 65.608 | 45.069 | 6.818 | 11.17 | 52.862 | 89.949 |
| 04 | 0.88 | 31.274 | 1.791 | 1.912 | 1.757 | 23.105 | 0.0 | 7.529 | 2.967 | 140.768 | 5.336 | 34.524 | 3.503 | 151.698 | 3.949 | 25.908 |
| 05 | 0.0 | 0.85 | 0.0 | 8.257 | 0.979 | 14.116 | 0.0 | 331.75 | 0.0 | 0.0 | 0.0 | 11.076 | 0.0 | 1.934 | 0.0 | 67.94 |
| 06 | 0.979 | 0.0 | 0.0 | 0.0 | 12.95 | 1.997 | 0.0 | 1.953 | 0.0 | 0.0 | 0.0 | 0.0 | 400.149 | 1.287 | 14.501 | 3.085 |
| 07 | 21.904 | 5.017 | 0.0 | 387.156 | 1.287 | 1.012 | 0.0 | 0.985 | 0.866 | 2.807 | 0.894 | 9.934 | 0.0 | 0.0 | 1.057 | 3.98 |
| 08 | 8.049 | 0.0 | 0.0 | 16.008 | 0.868 | 0.0 | 0.0 | 7.968 | 0.0 | 0.0 | 0.0 | 1.951 | 22.906 | 2.722 | 5.123 | 371.305 |
| 09 | 0.0 | 0.0 | 0.0 | 31.823 | 0.0 | 0.0 | 0.0 | 2.722 | 0.99 | 1.033 | 0.0 | 3.1 | 12.135 | 19.211 | 0.0 | 365.886 |
| 10 | 11.093 | 1.042 | 0.99 | 0.0 | 1.994 | 1.853 | 0.0 | 16.397 | 0.0 | 0.0 | 0.0 | 0.0 | 72.549 | 49.218 | 3.611 | 278.154 |
| 11 | 6.819 | 2.78 | 0.91 | 75.126 | 8.214 | 5.672 | 5.023 | 33.203 | 0.877 | 1.055 | 1.815 | 0.854 | 22.783 | 6.009 | 32.328 | 233.433 |
| 12 | 10.644 | 0.0 | 28.049 | 0.0 | 12.657 | 1.124 | 1.735 | 0.0 | 3.072 | 0.891 | 36.113 | 0.0 | 232.171 | 1.077 | 109.368 | 0.0 |
| 13 | 32.151 | 16.57 | 193.608 | 16.216 | 1.077 | 0.891 | 0.0 | 1.124 | 12.303 | 13.141 | 124.551 | 25.27 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 18.958 | 24.654 | 1.918 | 0.0 | 13.675 | 8.481 | 0.0 | 8.444 | 122.352 | 124.908 | 35.248 | 35.65 | 10.425 | 20.883 | 3.769 | 7.534 |
| 15 | 60.319 | 29.615 | 32.761 | 42.717 | 45.052 | 52.332 | 11.989 | 69.554 | 6.138 | 11.599 | 9.036 | 14.163 | 14.82 | 11.049 | 8.302 | 17.457 |
| 16 | 34.854 | 38.506 | 21.23 | 31.74 | 23.339 | 29.987 | 4.998 | 46.269 | 14.276 | 22.847 | 16.533 | 8.43 | 19.275 | 56.175 | 29.294 | 39.148 |
| 17 | 19.832 | 25.09 | 27.745 | 19.078 | 50.915 | 26.262 | 10.527 | 59.811 | 9.404 | 18.241 | 20.918 | 23.492 | 23.367 | 28.924 | 27.665 | 45.63 |
| 18 | 23.497 | 19.368 | 28.861 | 31.792 | 35.823 | 20.718 | 4.546 | 37.429 | 13.903 | 16.278 | 27.751 | 28.924 | 10.466 | 25.934 | 68.066 | 43.546 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.268 | -0.816 | 0.415 | -0.071 | -0.004 | -0.99 | -1.503 | 0.075 | 0.151 | 0.215 | 0.206 | 0.285 | -0.231 | -0.861 | 0.807 | -0.332 |
| 02 | 0.128 | -1.04 | 0.381 | 0.266 | -0.018 | -1.842 | -1.049 | -0.015 | 0.551 | -0.963 | 0.426 | 0.444 | -0.833 | -1.62 | 0.345 | 0.7 |
| 03 | -0.572 | -1.324 | 0.754 | 0.287 | -1.666 | -1.927 | -1.368 | -0.688 | -1.089 | -0.926 | 0.869 | 0.496 | -1.347 | -0.874 | 0.654 | 1.183 |
| 04 | -3.09 | 0.134 | -2.546 | -2.491 | -2.561 | -0.165 | -4.289 | -1.253 | -2.113 | 1.629 | -1.578 | 0.232 | -1.964 | 1.703 | -1.856 | -0.052 |
| 05 | -4.289 | -3.114 | -4.289 | -1.165 | -3.014 | -0.647 | -4.289 | 2.485 | -4.289 | -4.289 | -4.289 | -0.882 | -4.289 | -2.482 | -4.289 | 0.903 |
| 06 | -3.014 | -4.289 | -4.289 | -4.289 | -0.731 | -2.455 | -4.289 | -2.474 | -4.289 | -4.289 | -4.289 | -4.289 | 2.672 | -2.81 | -0.621 | -2.078 |
| 07 | -0.217 | -1.635 | -4.289 | 2.639 | -2.81 | -2.99 | -4.289 | -3.01 | -3.101 | -2.162 | -3.079 | -0.987 | -4.289 | -4.289 | -2.958 | -1.848 |
| 08 | -1.189 | -4.289 | -4.289 | -0.524 | -3.099 | -4.289 | -4.289 | -1.199 | -4.289 | -4.289 | -4.289 | -2.474 | -0.173 | -2.189 | -1.616 | 2.597 |
| 09 | -4.289 | -4.289 | -4.289 | 0.151 | -4.289 | -4.289 | -4.289 | -2.189 | -3.006 | -2.975 | -4.289 | -2.074 | -0.794 | -0.346 | -4.289 | 2.582 |
| 10 | -0.881 | -2.969 | -3.006 | -4.289 | -2.456 | -2.518 | -4.289 | -0.501 | -4.289 | -4.289 | -4.289 | -4.289 | 0.969 | 0.583 | -1.937 | 2.309 |
| 11 | -1.347 | -2.17 | -3.066 | 1.003 | -1.17 | -1.521 | -1.634 | 0.193 | -3.092 | -2.96 | -2.535 | -3.111 | -0.178 | -1.466 | 0.167 | 2.134 |
| 12 | -0.921 | -4.289 | 0.026 | -4.289 | -0.753 | -2.913 | -2.572 | -4.289 | -2.082 | -3.081 | 0.276 | -4.289 | 2.128 | -2.945 | 1.377 | -4.289 |
| 13 | 0.161 | -0.491 | 1.947 | -0.512 | -2.945 | -3.081 | -4.289 | -2.913 | -0.781 | -0.717 | 1.507 | -0.076 | -4.289 | -4.289 | -4.289 | -4.289 |
| 14 | -0.359 | -0.101 | -2.489 | -4.289 | -0.678 | -1.139 | -4.289 | -1.143 | 1.489 | 1.51 | 0.252 | 0.263 | -0.941 | -0.264 | -1.898 | -1.252 |
| 15 | 0.785 | 0.08 | 0.18 | 0.443 | 0.495 | 0.644 | -0.806 | 0.927 | -1.446 | -0.838 | -1.079 | -0.644 | -0.6 | -0.885 | -1.16 | -0.44 |
| 16 | 0.241 | 0.34 | -0.248 | 0.149 | -0.155 | 0.092 | -1.639 | 0.522 | -0.636 | -0.176 | -0.493 | -1.145 | -0.343 | 0.714 | 0.069 | 0.356 |
| 17 | -0.315 | -0.083 | 0.016 | -0.353 | 0.617 | -0.038 | -0.932 | 0.777 | -1.04 | -0.397 | -0.262 | -0.148 | -0.153 | 0.057 | 0.013 | 0.508 |
| 18 | -0.148 | -0.338 | 0.055 | 0.15 | 0.268 | -0.272 | -1.726 | 0.312 | -0.662 | -0.508 | 0.016 | 0.057 | -0.937 | -0.051 | 0.905 | 0.462 |