Model info
| Transcription factor | IRF7 | ||||||||
| Model | IRF7_HUMAN.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 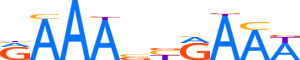 | ||||||||
| LOGO (reverse complement) | 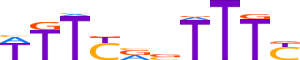 | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 10 | ||||||||
| Quality | C | ||||||||
| Consensus | RAAAbYRAAW | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 40 | ||||||||
| TF family | Interferon-regulatory factors{3.5.3} | ||||||||
| TF subfamily | IRF-7{3.5.3.0.7} | ||||||||
| HGNC | 6122 | ||||||||
| EntrezGene | 3665 | ||||||||
| UniProt ID | IRF7_HUMAN | ||||||||
| UniProt AC | Q92985 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 8.575 | 2.556 | 26.294 | 2.442 |
| 02 | 35.778 | 4.089 | 0.0 | 0.0 |
| 03 | 39.867 | 0.0 | 0.0 | 0.0 |
| 04 | 35.778 | 1.022 | 0.0 | 3.067 |
| 05 | 4.089 | 14.084 | 16.583 | 5.111 |
| 06 | 0.909 | 16.015 | 6.133 | 16.81 |
| 07 | 9.768 | 1.931 | 27.146 | 1.022 |
| 08 | 36.8 | 0.0 | 1.022 | 2.044 |
| 09 | 31.689 | 8.178 | 0.0 | 0.0 |
| 10 | 26.691 | 0.0 | 3.067 | 10.109 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.137 | -1.142 | 0.916 | -1.175 |
| 02 | 1.215 | -0.776 | -2.47 | -2.47 |
| 03 | 1.321 | -2.47 | -2.47 | -2.47 |
| 04 | 1.215 | -1.723 | -2.47 | -1.004 |
| 05 | -0.776 | 0.321 | 0.475 | -0.591 |
| 06 | -1.783 | 0.442 | -0.434 | 0.488 |
| 07 | -0.018 | -1.34 | 0.947 | -1.723 |
| 08 | 1.243 | -2.47 | -1.723 | -1.301 |
| 09 | 1.097 | -0.179 | -2.47 | -2.47 |
| 10 | 0.931 | -2.47 | -1.004 | 0.013 |