Model info
| Transcription factor | TCF12 | ||||||||
| Model | HTF4_HUMAN.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 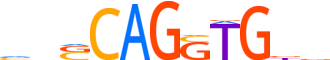 | ||||||||
| LOGO (reverse complement) | 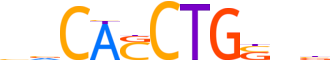 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 11 | ||||||||
| Quality | B | ||||||||
| Consensus | vnSCAGSTGbb | ||||||||
| wAUC | 0.7923845357850936 | ||||||||
| Best AUC | 0.8030591245829948 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 504 | ||||||||
| TF family | E2A-related factors{1.2.1} | ||||||||
| TF subfamily | HTF-4 (TCF-12, HEB){1.2.1.0.3} | ||||||||
| HGNC | 11623 | ||||||||
| EntrezGene | 6938 | ||||||||
| UniProt ID | HTF4_HUMAN | ||||||||
| UniProt AC | Q99081 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 133.137 | 183.886 | 138.615 | 44.362 |
| 02 | 132.719 | 88.533 | 156.963 | 121.785 |
| 03 | 69.774 | 246.667 | 182.562 | 0.997 |
| 04 | 5.965 | 485.927 | 2.803 | 5.306 |
| 05 | 493.806 | 0.925 | 3.489 | 1.78 |
| 06 | 1.169 | 0.0 | 498.831 | 0.0 |
| 07 | 25.408 | 114.649 | 350.702 | 9.241 |
| 08 | 40.597 | 5.885 | 12.456 | 441.062 |
| 09 | 0.0 | 4.498 | 488.959 | 6.543 |
| 10 | 36.281 | 152.42 | 141.467 | 169.833 |
| 11 | 76.097 | 104.956 | 213.078 | 105.87 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.062 | 0.382 | 0.102 | -1.014 |
| 02 | 0.059 | -0.34 | 0.225 | -0.026 |
| 03 | -0.573 | 0.674 | 0.375 | -3.904 |
| 04 | -2.823 | 1.349 | -3.369 | -2.915 |
| 05 | 1.365 | -3.933 | -3.223 | -3.637 |
| 06 | -3.839 | -4.4 | 1.375 | -4.4 |
| 07 | -1.546 | -0.085 | 1.024 | -2.462 |
| 08 | -1.099 | -2.834 | -2.201 | 1.252 |
| 09 | -4.4 | -3.04 | 1.355 | -2.749 |
| 10 | -1.207 | 0.196 | 0.122 | 0.303 |
| 11 | -0.488 | -0.172 | 0.528 | -0.164 |