Model info
| Transcription factor | GATA1 | ||||||||
| Model | GATA1_HUMAN.H10MO.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 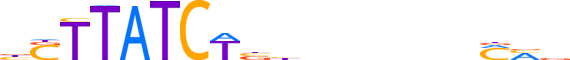 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | vbKnnnnnndvWGATAASv | ||||||||
| wAUC | 0.8861106951190332 | ||||||||
| Best AUC | 0.9333807602038442 | ||||||||
| Benchmark datasets | 12 | ||||||||
| Aligned words | 271 | ||||||||
| TF family | GATA-type zinc fingers{2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors{2.2.1.1} | ||||||||
| HGNC | 4170 | ||||||||
| EntrezGene | 2623 | ||||||||
| UniProt ID | GATA1_HUMAN | ||||||||
| UniProt AC | P15976 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 49.807 | 123.918 | 52.887 | 44.389 |
| 02 | 22.408 | 26.334 | 73.609 | 148.649 |
| 03 | 16.777 | 22.934 | 181.08 | 50.209 |
| 04 | 40.149 | 63.092 | 91.823 | 75.936 |
| 05 | 50.338 | 55.97 | 78.736 | 85.955 |
| 06 | 71.744 | 63.462 | 87.91 | 47.884 |
| 07 | 58.944 | 65.652 | 100.835 | 45.569 |
| 08 | 83.962 | 58.923 | 89.459 | 38.656 |
| 09 | 44.111 | 69.867 | 99.092 | 57.93 |
| 10 | 116.095 | 34.092 | 67.045 | 53.768 |
| 11 | 34.576 | 121.795 | 90.362 | 24.266 |
| 12 | 182.983 | 4.049 | 3.948 | 80.02 |
| 13 | 0.0 | 0.0 | 271.0 | 0.0 |
| 14 | 269.832 | 0.0 | 0.0 | 1.168 |
| 15 | 2.713 | 0.929 | 1.921 | 265.437 |
| 16 | 264.07 | 0.0 | 0.0 | 6.93 |
| 17 | 241.631 | 4.905 | 23.5 | 0.964 |
| 18 | 29.534 | 34.471 | 191.689 | 15.307 |
| 19 | 85.725 | 48.364 | 118.62 | 18.291 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.3 | 0.595 | -0.242 | -0.412 |
| 02 | -1.066 | -0.914 | 0.081 | 0.775 |
| 03 | -1.336 | -1.044 | 0.97 | -0.293 |
| 04 | -0.509 | -0.07 | 0.299 | 0.112 |
| 05 | -0.29 | -0.187 | 0.147 | 0.234 |
| 06 | 0.056 | -0.064 | 0.256 | -0.339 |
| 07 | -0.136 | -0.031 | 0.391 | -0.387 |
| 08 | 0.211 | -0.137 | 0.273 | -0.546 |
| 09 | -0.418 | 0.03 | 0.374 | -0.153 |
| 10 | 0.53 | -0.667 | -0.01 | -0.226 |
| 11 | -0.653 | 0.577 | 0.283 | -0.991 |
| 12 | 0.981 | -2.541 | -2.56 | 0.163 |
| 13 | -3.899 | -3.899 | 1.371 | -3.899 |
| 14 | 1.367 | -3.899 | -3.899 | -3.293 |
| 15 | -2.822 | -3.39 | -3.036 | 1.35 |
| 16 | 1.345 | -3.899 | -3.899 | -2.116 |
| 17 | 1.257 | -2.395 | -1.021 | -3.376 |
| 18 | -0.804 | -0.656 | 1.027 | -1.42 |
| 19 | 0.231 | -0.329 | 0.551 | -1.256 |