Model info
| Transcription factor | FLI1 | ||||||||
| Model | FLI1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 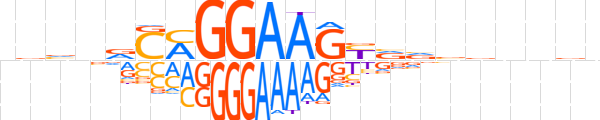 | ||||||||
| LOGO (reverse complement) | 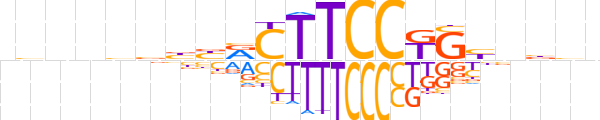 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Consensus | nvvnvCMGGAAGYvvbbnnvn | ||||||||
| wAUC | 0.9042392917898996 | ||||||||
| Best AUC | 0.940010483469558 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 522 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Ets-like factors{3.5.2.1} | ||||||||
| HGNC | 3749 | ||||||||
| EntrezGene | 2313 | ||||||||
| UniProt ID | FLI1_HUMAN | ||||||||
| UniProt AC | Q01543 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.341 | 31.578 | 45.561 | 16.628 | 42.51 | 37.184 | 40.469 | 23.898 | 27.866 | 51.891 | 77.867 | 12.45 | 4.099 | 23.133 | 30.659 | 17.865 |
| 02 | 10.722 | 32.275 | 37.505 | 10.315 | 53.033 | 55.104 | 23.675 | 11.974 | 60.001 | 71.692 | 44.953 | 17.91 | 11.694 | 26.486 | 26.912 | 5.749 |
| 03 | 30.69 | 31.786 | 65.24 | 7.732 | 48.336 | 59.279 | 22.654 | 55.289 | 27.827 | 33.685 | 66.62 | 4.914 | 5.953 | 10.956 | 23.175 | 5.863 |
| 04 | 63.956 | 6.733 | 36.337 | 5.78 | 64.542 | 16.791 | 37.496 | 16.877 | 74.094 | 6.65 | 90.948 | 5.997 | 18.069 | 12.071 | 35.644 | 8.014 |
| 05 | 3.166 | 169.128 | 48.368 | 0.0 | 0.0 | 42.245 | 0.0 | 0.0 | 13.673 | 152.621 | 34.132 | 0.0 | 0.0 | 31.577 | 5.092 | 0.0 |
| 06 | 15.978 | 0.861 | 0.0 | 0.0 | 213.127 | 181.481 | 0.963 | 0.0 | 48.451 | 39.14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 277.556 | 0.0 | 0.0 | 0.0 | 221.481 | 0.0 | 0.0 | 0.0 | 0.963 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 500.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 495.136 | 2.81 | 1.162 | 0.893 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 458.855 | 0.0 | 1.901 | 34.379 | 0.97 | 0.0 | 0.947 | 0.893 | 1.162 | 0.0 | 0.0 | 0.0 | 0.893 | 0.0 | 0.0 | 0.0 |
| 11 | 90.193 | 5.033 | 366.653 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.848 | 0.0 | 1.904 | 0.0 | 33.368 | 0.0 |
| 12 | 2.762 | 40.082 | 8.876 | 40.377 | 0.0 | 4.121 | 0.0 | 0.912 | 4.947 | 120.617 | 48.688 | 228.618 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 0.0 | 1.901 | 5.808 | 0.0 | 67.567 | 29.386 | 51.716 | 16.151 | 26.09 | 8.384 | 20.368 | 2.722 | 15.016 | 43.15 | 199.195 | 12.547 |
| 14 | 24.449 | 19.455 | 54.861 | 9.908 | 25.538 | 19.535 | 21.696 | 16.052 | 75.488 | 49.567 | 138.203 | 13.829 | 3.144 | 3.99 | 24.285 | 0.0 |
| 15 | 13.4 | 54.682 | 46.359 | 14.178 | 16.577 | 38.114 | 17.434 | 20.421 | 39.587 | 86.691 | 75.703 | 37.066 | 2.812 | 11.784 | 19.34 | 5.853 |
| 16 | 20.093 | 10.175 | 35.115 | 6.993 | 32.99 | 35.18 | 65.594 | 57.506 | 38.178 | 38.052 | 62.823 | 19.783 | 2.923 | 11.76 | 47.111 | 15.724 |
| 17 | 16.553 | 18.49 | 52.413 | 6.728 | 29.734 | 15.803 | 17.198 | 32.431 | 43.105 | 60.831 | 66.748 | 39.959 | 4.804 | 34.907 | 40.479 | 19.816 |
| 18 | 21.434 | 29.906 | 32.996 | 9.86 | 34.498 | 32.713 | 30.986 | 31.834 | 30.444 | 49.964 | 66.306 | 30.124 | 10.583 | 29.811 | 41.736 | 16.805 |
| 19 | 9.621 | 15.878 | 55.958 | 15.501 | 36.456 | 39.303 | 35.239 | 31.397 | 35.564 | 52.694 | 62.954 | 20.812 | 9.829 | 20.821 | 44.186 | 13.786 |
| 20 | 19.13 | 22.315 | 38.263 | 11.762 | 34.447 | 29.581 | 17.046 | 47.622 | 48.576 | 62.748 | 65.467 | 21.548 | 5.783 | 25.909 | 34.436 | 15.37 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.637 | 0.01 | 0.373 | -0.62 | 0.304 | 0.172 | 0.256 | -0.264 | -0.113 | 0.502 | 0.906 | -0.902 | -1.953 | -0.296 | -0.019 | -0.55 |
| 02 | -1.047 | 0.032 | 0.18 | -1.084 | 0.524 | 0.562 | -0.274 | -0.94 | 0.646 | 0.823 | 0.36 | -0.548 | -0.963 | -0.163 | -0.147 | -1.64 |
| 03 | -0.018 | 0.017 | 0.73 | -1.36 | 0.432 | 0.634 | -0.317 | 0.565 | -0.115 | 0.074 | 0.75 | -1.786 | -1.607 | -1.026 | -0.295 | -1.622 |
| 04 | 0.71 | -1.491 | 0.149 | -1.635 | 0.719 | -0.611 | 0.18 | -0.606 | 0.856 | -1.503 | 1.06 | -1.6 | -0.539 | -0.932 | 0.13 | -1.326 |
| 05 | -2.186 | 1.679 | 0.432 | -4.4 | -4.4 | 0.298 | -4.4 | -4.4 | -0.811 | 1.576 | 0.087 | -4.4 | -4.4 | 0.01 | -1.753 | -4.4 |
| 06 | -0.659 | -3.232 | -4.4 | -4.4 | 1.909 | 1.749 | -3.153 | -4.4 | 0.434 | 0.223 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | 2.173 | -4.4 | -4.4 | -4.4 | 1.948 | -4.4 | -4.4 | -4.4 | -3.153 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.761 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.751 | -2.292 | -3.016 | -3.207 | -4.4 | -4.4 | -4.4 | -4.4 |
| 10 | 2.675 | -4.4 | -2.626 | 0.094 | -3.148 | -4.4 | -3.165 | -3.207 | -3.016 | -4.4 | -4.4 | -4.4 | -3.207 | -4.4 | -4.4 | -4.4 |
| 11 | 1.052 | -1.764 | 2.451 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.28 | -4.4 | -2.625 | -4.4 | 0.065 | -4.4 |
| 12 | -2.307 | 0.246 | -1.228 | 0.253 | -4.4 | -1.948 | -4.4 | -3.192 | -1.78 | 1.341 | 0.439 | 1.979 | -4.4 | -4.4 | -4.4 | -4.4 |
| 13 | -4.4 | -2.626 | -1.63 | -4.4 | 0.764 | -0.061 | 0.499 | -0.649 | -0.178 | -1.283 | -0.422 | -2.32 | -0.72 | 0.319 | 1.842 | -0.894 |
| 14 | -0.242 | -0.467 | 0.557 | -1.123 | -0.199 | -0.462 | -0.36 | -0.655 | 0.875 | 0.457 | 1.477 | -0.8 | -2.192 | -1.978 | -0.249 | -4.4 |
| 15 | -0.831 | 0.554 | 0.39 | -0.776 | -0.623 | 0.196 | -0.574 | -0.419 | 0.234 | 1.012 | 0.878 | 0.169 | -2.291 | -0.955 | -0.472 | -1.623 |
| 16 | -0.435 | -1.097 | 0.115 | -1.455 | 0.054 | 0.117 | 0.735 | 0.604 | 0.198 | 0.195 | 0.692 | -0.45 | -2.257 | -0.957 | 0.406 | -0.675 |
| 17 | -0.625 | -0.516 | 0.512 | -1.492 | -0.049 | -0.67 | -0.587 | 0.037 | 0.318 | 0.66 | 0.752 | 0.243 | -1.807 | 0.109 | 0.256 | -0.448 |
| 18 | -0.371 | -0.043 | 0.054 | -1.127 | 0.098 | 0.045 | -0.008 | 0.018 | -0.026 | 0.465 | 0.746 | -0.036 | -1.059 | -0.047 | 0.286 | -0.61 |
| 19 | -1.151 | -0.665 | 0.577 | -0.689 | 0.152 | 0.227 | 0.119 | 0.005 | 0.128 | 0.517 | 0.694 | -0.4 | -1.13 | -0.4 | 0.343 | -0.803 |
| 20 | -0.483 | -0.332 | 0.2 | -0.957 | 0.096 | -0.054 | -0.596 | 0.417 | 0.437 | 0.691 | 0.733 | -0.366 | -1.634 | -0.185 | 0.096 | -0.697 |