Model info
| Transcription factor | BARHL2 | ||||||||
| Model | BARH2_HUMAN.H10MO.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 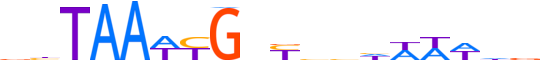 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HT-SELEX | ||||||||
| Model length | 18 | ||||||||
| Quality | D | ||||||||
| Consensus | vhTAAWYGnYbhhWWWhd | ||||||||
| wAUC | |||||||||
| Best AUC | |||||||||
| Benchmark datasets | |||||||||
| Aligned words | 41168 | ||||||||
| TF family | NK-related factors{3.1.2} | ||||||||
| TF subfamily | BARHL{3.1.2.1} | ||||||||
| HGNC | 954 | ||||||||
| EntrezGene | 343472 | ||||||||
| UniProt ID | BARH2_HUMAN | ||||||||
| UniProt AC | Q9NY43 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 9476.249 | 9833.782 | 17728.772 | 5786.443 |
| 02 | 7154.271 | 16490.133 | 5197.05 | 13983.793 |
| 03 | 386.423 | 266.412 | 259.449 | 41912.963 |
| 04 | 42328.52 | 27.852 | 63.3 | 405.574 |
| 05 | 42751.66 | 33.391 | 27.694 | 12.502 |
| 06 | 20391.88 | 77.384 | 1322.659 | 21033.324 |
| 07 | 708.895 | 19177.091 | 722.708 | 22216.552 |
| 08 | 902.695 | 72.377 | 41600.183 | 249.991 |
| 09 | 8744.555 | 10605.559 | 14843.186 | 8631.947 |
| 10 | 2423.369 | 11796.264 | 2673.031 | 25932.583 |
| 11 | 6191.882 | 16422.315 | 12607.007 | 7604.042 |
| 12 | 9365.508 | 16247.701 | 5720.577 | 11491.46 |
| 13 | 11500.954 | 5238.928 | 4330.084 | 21755.28 |
| 14 | 22377.751 | 892.482 | 2133.519 | 17421.495 |
| 15 | 21000.289 | 791.372 | 587.952 | 20445.633 |
| 16 | 17982.745 | 1379.672 | 2126.954 | 21335.875 |
| 17 | 14410.464 | 7798.389 | 5053.833 | 15562.56 |
| 18 | 9772.326 | 6668.139 | 18648.688 | 7736.093 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.122 | -0.085 | 0.504 | -0.615 |
| 02 | -0.403 | 0.432 | -0.722 | 0.267 |
| 03 | -3.315 | -3.684 | -3.71 | 1.365 |
| 04 | 1.374 | -5.861 | -5.09 | -3.267 |
| 05 | 1.384 | -5.694 | -5.866 | -6.56 |
| 06 | 0.644 | -4.896 | -2.089 | 0.675 |
| 07 | -2.711 | 0.583 | -2.692 | 0.73 |
| 08 | -2.471 | -4.961 | 1.357 | -3.747 |
| 09 | -0.202 | -0.009 | 0.327 | -0.215 |
| 10 | -1.485 | 0.097 | -1.387 | 0.885 |
| 11 | -0.547 | 0.428 | 0.163 | -0.342 |
| 12 | -0.134 | 0.417 | -0.627 | 0.071 |
| 13 | 0.072 | -0.714 | -0.905 | 0.709 |
| 14 | 0.737 | -2.482 | -1.612 | 0.487 |
| 15 | 0.674 | -2.602 | -2.898 | 0.647 |
| 16 | 0.518 | -2.047 | -1.615 | 0.689 |
| 17 | 0.297 | -0.317 | -0.75 | 0.374 |
| 18 | -0.091 | -0.473 | 0.555 | -0.325 |