Model info
| Transcription factor | ATF3 | ||||||||
| Model | ATF3_HUMAN.H10MO.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 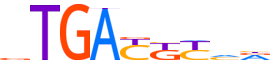 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HOCOMOCO v9 | ||||||||
| Model length | 9 | ||||||||
| Quality | A | ||||||||
| Consensus | vTGAYKYvd | ||||||||
| wAUC | 0.7038527078022759 | ||||||||
| Best AUC | 0.7038527078022759 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 2096 | ||||||||
| TF family | Fos-related factors{1.1.2} | ||||||||
| TF subfamily | ATF-3-like factors{1.1.2.2} | ||||||||
| HGNC | 785 | ||||||||
| EntrezGene | 467 | ||||||||
| UniProt ID | ATF3_HUMAN | ||||||||
| UniProt AC | P18847 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 453.184 | 431.487 | 881.269 | 270.06 |
| 02 | 14.5 | 8.957 | 12.241 | 2000.302 |
| 03 | 2.995 | 17.967 | 2010.822 | 4.216 |
| 04 | 1971.866 | 14.842 | 9.194 | 40.098 |
| 05 | 13.16 | 1319.297 | 137.619 | 565.924 |
| 06 | 54.039 | 129.41 | 1120.827 | 731.723 |
| 07 | 109.39 | 1023.77 | 26.058 | 876.782 |
| 08 | 679.979 | 892.398 | 258.527 | 205.095 |
| 09 | 1139.729 | 255.965 | 323.162 | 317.144 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.116 | -0.165 | 0.547 | -0.631 |
| 02 | -3.439 | -3.851 | -3.587 | 1.366 |
| 03 | -4.647 | -3.247 | 1.371 | -4.425 |
| 04 | 1.352 | -3.418 | -3.829 | -2.498 |
| 05 | -3.524 | 0.95 | -1.298 | 0.106 |
| 06 | -2.212 | -1.359 | 0.787 | 0.362 |
| 07 | -1.524 | 0.697 | -2.905 | 0.542 |
| 08 | 0.289 | 0.56 | -0.674 | -0.903 |
| 09 | 0.804 | -0.684 | -0.452 | -0.471 |