Model info
| Transcription factor | ATF1 | ||||||||
| Model | ATF1_HUMAN.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 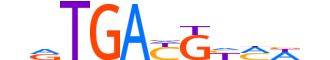 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 11 | ||||||||
| Quality | B | ||||||||
| Consensus | nvTGAYGYMWb | ||||||||
| wAUC | 0.7969790617507443 | ||||||||
| Best AUC | 0.8550480218463526 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | CREB-related factors{1.1.7} | ||||||||
| TF subfamily | CREB-like factors{1.1.7.1} | ||||||||
| HGNC | 783 | ||||||||
| EntrezGene | 466 | ||||||||
| UniProt ID | ATF1_HUMAN | ||||||||
| UniProt AC | P18846 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 123.799 | 89.468 | 144.178 | 142.555 |
| 02 | 195.235 | 39.317 | 231.474 | 33.974 |
| 03 | 0.925 | 0.0 | 0.0 | 499.075 |
| 04 | 2.941 | 0.0 | 492.551 | 4.508 |
| 05 | 492.539 | 0.845 | 5.794 | 0.823 |
| 06 | 41.084 | 334.641 | 27.29 | 96.985 |
| 07 | 0.0 | 3.027 | 370.984 | 125.989 |
| 08 | 72.39 | 121.298 | 24.727 | 281.585 |
| 09 | 194.208 | 247.205 | 18.068 | 40.519 |
| 10 | 299.506 | 43.614 | 55.637 | 101.243 |
| 11 | 84.076 | 85.897 | 143.044 | 186.983 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.01 | -0.33 | 0.141 | 0.13 |
| 02 | 0.441 | -1.13 | 0.61 | -1.27 |
| 03 | -3.933 | -4.4 | -4.4 | 1.375 |
| 04 | -3.338 | -4.4 | 1.362 | -3.039 |
| 05 | 1.362 | -3.966 | -2.846 | -3.975 |
| 06 | -1.088 | 0.977 | -1.479 | -0.25 |
| 07 | -4.4 | -3.319 | 1.08 | 0.008 |
| 08 | -0.537 | -0.03 | -1.572 | 0.805 |
| 09 | 0.436 | 0.676 | -1.864 | -1.101 |
| 10 | 0.867 | -1.03 | -0.794 | -0.208 |
| 11 | -0.391 | -0.37 | 0.133 | 0.399 |