Model info
| Transcription factor | AR | ||||||||
| Model | ANDR_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 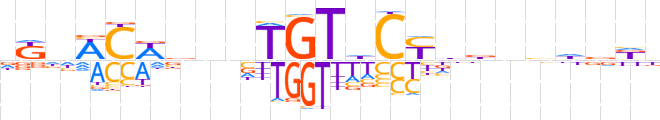 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | dKnWCWbnvTGThCYbbnbhddd | ||||||||
| wAUC | 0.8695772129164118 | ||||||||
| Best AUC | 0.9139229205204544 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 498 | ||||||||
| TF family | Steroid hormone receptors (NR3){2.1.1} | ||||||||
| TF subfamily | GR-like receptors (NR3C){2.1.1.1} | ||||||||
| HGNC | 644 | ||||||||
| EntrezGene | 367 | ||||||||
| UniProt ID | ANDR_HUMAN | ||||||||
| UniProt AC | P10275 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 15.447 | 4.85 | 117.394 | 30.532 | 10.03 | 1.967 | 8.157 | 11.194 | 20.876 | 3.11 | 119.883 | 25.866 | 12.641 | 3.859 | 82.091 | 19.164 |
| 02 | 21.86 | 8.441 | 9.827 | 18.867 | 8.051 | 1.96 | 0.0 | 3.776 | 101.74 | 64.516 | 60.122 | 101.148 | 15.487 | 19.351 | 12.984 | 38.934 |
| 03 | 110.167 | 10.318 | 15.787 | 10.867 | 80.281 | 0.988 | 1.022 | 11.976 | 55.283 | 12.622 | 5.753 | 9.274 | 110.082 | 11.282 | 11.948 | 29.413 |
| 04 | 0.0 | 312.835 | 20.825 | 22.153 | 0.947 | 29.141 | 0.0 | 5.122 | 2.193 | 27.332 | 0.0 | 4.985 | 1.988 | 49.624 | 3.16 | 6.759 |
| 05 | 0.956 | 0.0 | 1.969 | 2.203 | 278.878 | 11.612 | 23.726 | 104.716 | 8.138 | 1.923 | 7.795 | 6.129 | 11.096 | 1.955 | 7.906 | 18.062 |
| 06 | 35.301 | 85.87 | 118.911 | 58.986 | 2.921 | 8.781 | 0.964 | 2.824 | 8.95 | 9.853 | 6.89 | 15.703 | 13.238 | 25.265 | 56.7 | 35.907 |
| 07 | 18.878 | 4.907 | 21.683 | 14.942 | 50.443 | 22.979 | 7.925 | 48.422 | 67.301 | 30.009 | 36.907 | 49.248 | 16.97 | 21.691 | 38.003 | 36.755 |
| 08 | 40.7 | 60.132 | 39.703 | 13.056 | 19.917 | 26.551 | 16.846 | 16.272 | 22.786 | 46.721 | 23.118 | 11.894 | 31.186 | 41.763 | 53.932 | 22.486 |
| 09 | 5.044 | 5.489 | 1.05 | 103.005 | 21.634 | 8.13 | 0.0 | 145.403 | 7.953 | 9.062 | 4.095 | 112.49 | 1.963 | 1.014 | 0.918 | 59.811 |
| 10 | 1.089 | 4.002 | 30.563 | 0.94 | 4.046 | 7.27 | 11.445 | 0.935 | 0.0 | 3.042 | 3.021 | 0.0 | 5.977 | 7.89 | 404.006 | 2.837 |
| 11 | 0.0 | 1.004 | 0.0 | 10.107 | 0.0 | 0.0 | 0.923 | 21.281 | 6.938 | 6.684 | 0.0 | 435.414 | 0.935 | 0.0 | 0.0 | 3.777 |
| 12 | 0.953 | 2.039 | 0.934 | 3.947 | 3.775 | 1.004 | 0.0 | 2.909 | 0.0 | 0.0 | 0.0 | 0.923 | 97.934 | 72.725 | 66.787 | 233.135 |
| 13 | 3.975 | 96.772 | 0.891 | 1.024 | 0.0 | 70.712 | 0.0 | 5.056 | 1.971 | 64.753 | 0.0 | 0.996 | 4.376 | 223.435 | 0.935 | 12.169 |
| 14 | 0.0 | 1.933 | 0.0 | 8.389 | 22.58 | 195.073 | 0.0 | 238.017 | 0.0 | 0.0 | 0.0 | 1.825 | 0.0 | 9.624 | 0.93 | 8.691 |
| 15 | 6.853 | 2.043 | 4.945 | 8.738 | 61.127 | 39.973 | 5.047 | 100.483 | 0.0 | 0.93 | 0.0 | 0.0 | 26.115 | 52.16 | 90.786 | 87.862 |
| 16 | 8.908 | 6.835 | 50.684 | 27.668 | 34.215 | 7.087 | 3.157 | 50.648 | 5.804 | 16.378 | 25.61 | 52.987 | 13.93 | 34.831 | 68.376 | 79.947 |
| 17 | 16.902 | 8.839 | 13.652 | 23.463 | 28.288 | 15.144 | 0.979 | 20.72 | 22.912 | 25.818 | 63.724 | 35.373 | 31.287 | 41.51 | 54.157 | 84.295 |
| 18 | 28.388 | 40.606 | 17.672 | 12.723 | 19.595 | 35.574 | 0.934 | 35.208 | 17.165 | 61.291 | 25.379 | 28.678 | 17.642 | 58.246 | 39.592 | 48.371 |
| 19 | 27.066 | 13.536 | 13.92 | 28.268 | 58.424 | 18.916 | 5.977 | 112.401 | 18.128 | 18.056 | 4.991 | 42.402 | 21.998 | 16.788 | 34.579 | 51.614 |
| 20 | 33.499 | 32.176 | 45.575 | 14.365 | 29.296 | 4.949 | 2.032 | 31.018 | 10.991 | 9.898 | 18.966 | 19.611 | 39.696 | 24.914 | 146.939 | 23.136 |
| 21 | 14.401 | 13.756 | 26.393 | 58.933 | 26.284 | 6.718 | 1.952 | 36.984 | 18.048 | 17.507 | 21.274 | 156.684 | 7.981 | 9.855 | 38.964 | 31.332 |
| 22 | 13.451 | 8.971 | 31.147 | 13.145 | 19.347 | 9.064 | 1.122 | 18.302 | 13.049 | 27.009 | 14.114 | 34.409 | 36.74 | 28.559 | 58.411 | 160.223 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.666 | -1.773 | 1.34 | 0.003 | -1.085 | -2.572 | -1.283 | -0.979 | -0.371 | -2.177 | 1.361 | -0.161 | -0.861 | -1.982 | 0.984 | -0.455 |
| 02 | -0.326 | -1.251 | -1.105 | -0.471 | -1.296 | -2.575 | -4.378 | -2.002 | 1.198 | 0.744 | 0.674 | 1.192 | -0.664 | -0.446 | -0.835 | 0.243 |
| 03 | 1.277 | -1.058 | -0.645 | -1.008 | 0.962 | -3.11 | -3.086 | -0.914 | 0.591 | -0.863 | -1.614 | -1.16 | 1.276 | -0.972 | -0.916 | -0.034 |
| 04 | -4.378 | 2.318 | -0.374 | -0.313 | -3.14 | -0.043 | -4.378 | -1.722 | -2.481 | -0.106 | -4.378 | -1.747 | -2.564 | 0.484 | -2.162 | -1.462 |
| 05 | -3.134 | -4.378 | -2.572 | -2.477 | 2.204 | -0.944 | -0.246 | 1.227 | -1.285 | -2.591 | -1.327 | -1.554 | -0.988 | -2.578 | -1.313 | -0.513 |
| 06 | 0.146 | 1.029 | 1.353 | 0.655 | -2.232 | -1.213 | -3.128 | -2.262 | -1.195 | -1.102 | -1.444 | -0.65 | -0.817 | -0.184 | 0.616 | 0.163 |
| 07 | -0.47 | -1.762 | -0.334 | -0.699 | 0.5 | -0.277 | -1.311 | 0.459 | 0.786 | -0.014 | 0.19 | 0.476 | -0.574 | -0.334 | 0.219 | 0.186 |
| 08 | 0.287 | 0.675 | 0.263 | -0.83 | -0.418 | -0.135 | -0.582 | -0.616 | -0.285 | 0.424 | -0.271 | -0.92 | 0.024 | 0.313 | 0.566 | -0.298 |
| 09 | -1.736 | -1.658 | -3.066 | 1.21 | -0.336 | -1.286 | -4.378 | 1.554 | -1.307 | -1.183 | -1.928 | 1.298 | -2.574 | -3.091 | -3.162 | 0.669 |
| 10 | -3.039 | -1.949 | 0.004 | -3.146 | -1.939 | -1.393 | -0.958 | -3.15 | -4.378 | -2.196 | -2.202 | -4.378 | -1.578 | -1.315 | 2.574 | -2.258 |
| 11 | -4.378 | -3.099 | -4.378 | -1.078 | -4.378 | -4.378 | -3.159 | -0.353 | -1.437 | -1.472 | -4.378 | 2.649 | -3.15 | -4.378 | -4.378 | -2.002 |
| 12 | -3.136 | -2.542 | -3.15 | -1.962 | -2.003 | -3.099 | -4.378 | -2.236 | -4.378 | -4.378 | -4.378 | -3.159 | 1.16 | 0.864 | 0.779 | 2.025 |
| 13 | -1.956 | 1.148 | -3.184 | -3.084 | -4.378 | 0.836 | -4.378 | -1.734 | -2.571 | 0.748 | -4.378 | -3.104 | -1.868 | 1.982 | -3.15 | -0.898 |
| 14 | -4.378 | -2.587 | -4.378 | -1.256 | -0.294 | 1.847 | -4.378 | 2.046 | -4.378 | -4.378 | -4.378 | -2.634 | -4.378 | -1.125 | -3.153 | -1.223 |
| 15 | -1.449 | -2.54 | -1.755 | -1.217 | 0.691 | 0.269 | -1.736 | 1.185 | -4.378 | -3.153 | -4.378 | -4.378 | -0.151 | 0.533 | 1.084 | 1.052 |
| 16 | -1.199 | -1.451 | 0.505 | -0.094 | 0.115 | -1.417 | -2.163 | 0.504 | -1.605 | -0.609 | -0.17 | 0.549 | -0.767 | 0.133 | 0.802 | 0.958 |
| 17 | -0.578 | -1.206 | -0.787 | -0.257 | -0.072 | -0.686 | -3.116 | -0.379 | -0.28 | -0.162 | 0.732 | 0.148 | 0.027 | 0.307 | 0.571 | 1.01 |
| 18 | -0.069 | 0.285 | -0.535 | -0.855 | -0.434 | 0.154 | -3.15 | 0.144 | -0.563 | 0.693 | -0.179 | -0.059 | -0.536 | 0.643 | 0.26 | 0.458 |
| 19 | -0.116 | -0.795 | -0.768 | -0.073 | 0.646 | -0.468 | -1.578 | 1.297 | -0.51 | -0.514 | -1.746 | 0.328 | -0.32 | -0.585 | 0.126 | 0.523 |
| 20 | 0.095 | 0.055 | 0.399 | -0.737 | -0.038 | -1.754 | -2.545 | 0.019 | -0.997 | -1.098 | -0.466 | -0.433 | 0.263 | -0.198 | 1.564 | -0.27 |
| 21 | -0.735 | -0.779 | -0.141 | 0.655 | -0.145 | -1.468 | -2.579 | 0.192 | -0.514 | -0.544 | -0.353 | 1.628 | -1.304 | -1.102 | 0.244 | 0.028 |
| 22 | -0.801 | -1.192 | 0.023 | -0.823 | -0.446 | -1.182 | -3.017 | -0.501 | -0.83 | -0.118 | -0.754 | 0.121 | 0.186 | -0.063 | 0.646 | 1.651 |