Model info
| Transcription factor | NFIC | ||||||||
| Model | NFIC_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 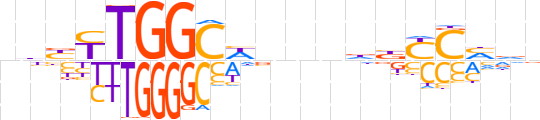 | ||||||||
| LOGO (reverse complement) | 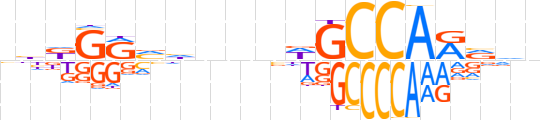 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | nhbYTGGCWbnndbYChdn | ||||||||
| wAUC | 0.8321439979573494 | ||||||||
| Best AUC | 0.8513505315960385 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | Nuclear factor 1{7.1.2} | ||||||||
| TF subfamily | NF-1C (NF-IC){7.1.2.0.3} | ||||||||
| HGNC | 7786 | ||||||||
| EntrezGene | 4782 | ||||||||
| UniProt ID | NFIC_HUMAN | ||||||||
| UniProt AC | P08651 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 36.264 | 15.323 | 25.388 | 41.837 | 47.739 | 22.372 | 2.916 | 51.537 | 22.338 | 36.146 | 25.058 | 31.692 | 26.914 | 27.07 | 15.663 | 71.744 |
| 02 | 9.31 | 53.616 | 37.148 | 33.18 | 3.949 | 68.444 | 3.308 | 25.21 | 1.054 | 42.508 | 11.49 | 13.973 | 9.092 | 96.008 | 22.161 | 69.549 |
| 03 | 0.0 | 9.448 | 0.0 | 13.956 | 0.0 | 112.521 | 2.264 | 145.791 | 0.0 | 53.692 | 0.0 | 20.416 | 0.0 | 35.6 | 2.854 | 103.458 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 0.902 | 0.0 | 1.969 | 208.39 | 0.0 | 1.132 | 0.0 | 3.986 | 0.0 | 0.0 | 8.759 | 274.862 |
| 05 | 0.0 | 0.0 | 0.902 | 0.0 | 0.0 | 0.0 | 1.132 | 0.0 | 0.874 | 0.0 | 9.854 | 0.0 | 0.928 | 0.952 | 485.357 | 0.0 |
| 06 | 0.0 | 0.0 | 1.802 | 0.0 | 0.0 | 0.0 | 0.952 | 0.0 | 1.103 | 0.843 | 493.267 | 2.032 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 1.103 | 0.0 | 0.843 | 0.0 | 0.0 | 0.0 | 48.085 | 436.225 | 9.1 | 2.612 | 0.0 | 0.996 | 1.036 | 0.0 |
| 08 | 23.389 | 1.017 | 7.329 | 17.194 | 234.818 | 57.765 | 9.809 | 134.83 | 4.124 | 1.036 | 2.134 | 3.945 | 0.0 | 0.0 | 0.0 | 2.612 |
| 09 | 47.738 | 64.382 | 114.61 | 35.6 | 20.231 | 23.566 | 3.265 | 12.755 | 2.755 | 8.051 | 2.981 | 5.485 | 5.543 | 43.494 | 67.974 | 41.569 |
| 10 | 25.792 | 14.5 | 18.667 | 17.308 | 49.465 | 30.866 | 12.216 | 46.946 | 51.423 | 58.047 | 41.14 | 38.22 | 13.763 | 31.762 | 28.961 | 20.922 |
| 11 | 32.984 | 23.936 | 69.144 | 14.38 | 39.517 | 60.052 | 6.828 | 28.778 | 33.197 | 18.237 | 37.012 | 12.539 | 13.93 | 26.08 | 61.934 | 21.452 |
| 12 | 41.293 | 5.618 | 35.161 | 37.557 | 55.594 | 12.538 | 4.261 | 55.913 | 65.131 | 14.028 | 54.367 | 41.392 | 11.851 | 7.382 | 21.115 | 36.8 |
| 13 | 11.642 | 27.053 | 89.681 | 45.492 | 4.465 | 4.363 | 8.602 | 22.135 | 11.222 | 19.579 | 62.147 | 21.957 | 7.784 | 17.172 | 85.126 | 61.581 |
| 14 | 8.843 | 15.345 | 0.902 | 10.023 | 7.495 | 38.401 | 1.881 | 20.39 | 21.783 | 198.309 | 4.445 | 21.019 | 11.201 | 104.282 | 2.955 | 32.728 |
| 15 | 7.51 | 33.473 | 8.338 | 0.0 | 7.269 | 329.859 | 0.0 | 19.209 | 0.81 | 3.609 | 5.764 | 0.0 | 5.781 | 55.513 | 14.348 | 8.518 |
| 16 | 4.679 | 9.333 | 6.431 | 0.928 | 222.558 | 151.832 | 7.514 | 40.55 | 7.81 | 8.815 | 3.269 | 8.555 | 1.863 | 14.641 | 4.814 | 6.407 |
| 17 | 85.636 | 37.51 | 93.952 | 19.813 | 113.979 | 14.426 | 9.496 | 46.718 | 9.088 | 5.199 | 5.972 | 1.769 | 5.687 | 6.408 | 30.734 | 13.611 |
| 18 | 40.562 | 24.596 | 107.599 | 41.634 | 21.859 | 16.466 | 4.009 | 21.21 | 42.372 | 38.287 | 30.72 | 28.774 | 13.247 | 21.799 | 36.608 | 10.258 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.147 | -0.7 | -0.205 | 0.289 | 0.419 | -0.329 | -2.259 | 0.495 | -0.331 | 0.144 | -0.218 | 0.014 | -0.147 | -0.142 | -0.679 | 0.824 |
| 02 | -1.182 | 0.535 | 0.171 | 0.059 | -1.987 | 0.777 | -2.147 | -0.212 | -3.088 | 0.304 | -0.98 | -0.79 | -1.205 | 1.114 | -0.339 | 0.793 |
| 03 | -4.4 | -1.168 | -4.4 | -0.791 | -4.4 | 1.272 | -2.479 | 1.53 | -4.4 | 0.536 | -4.4 | -0.419 | -4.4 | 0.129 | -2.278 | 1.189 |
| 04 | -4.4 | -4.4 | -4.4 | -4.4 | -3.199 | -4.4 | -2.597 | 1.887 | -4.4 | -3.035 | -4.4 | -1.979 | -4.4 | -4.4 | -1.241 | 2.163 |
| 05 | -4.4 | -4.4 | -3.199 | -4.4 | -4.4 | -4.4 | -3.035 | -4.4 | -3.221 | -4.4 | -1.128 | -4.4 | -3.179 | -3.161 | 2.731 | -4.4 |
| 06 | -4.4 | -4.4 | -2.67 | -4.4 | -4.4 | -4.4 | -3.161 | -4.4 | -3.055 | -3.246 | 2.747 | -2.57 | -4.4 | -4.4 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | -3.055 | -4.4 | -3.246 | -4.4 | -4.4 | -4.4 | 0.427 | 2.625 | -1.204 | -2.356 | -4.4 | -3.129 | -3.101 | -4.4 |
| 08 | -0.286 | -3.114 | -1.411 | -0.587 | 2.006 | 0.609 | -1.132 | 1.453 | -1.947 | -3.101 | -2.529 | -1.988 | -4.4 | -4.4 | -4.4 | -2.356 |
| 09 | 0.419 | 0.716 | 1.291 | 0.129 | -0.428 | -0.278 | -2.159 | -0.878 | -2.309 | -1.321 | -2.24 | -1.684 | -1.674 | 0.327 | 0.77 | 0.282 |
| 10 | -0.189 | -0.754 | -0.507 | -0.581 | 0.455 | -0.012 | -0.92 | 0.403 | 0.493 | 0.614 | 0.272 | 0.199 | -0.805 | 0.016 | -0.075 | -0.395 |
| 11 | 0.053 | -0.263 | 0.787 | -0.762 | 0.232 | 0.647 | -1.478 | -0.081 | 0.06 | -0.53 | 0.167 | -0.895 | -0.793 | -0.178 | 0.678 | -0.371 |
| 12 | 0.276 | -1.662 | 0.117 | 0.182 | 0.571 | -0.895 | -1.918 | 0.576 | 0.728 | -0.786 | 0.548 | 0.278 | -0.95 | -1.404 | -0.386 | 0.162 |
| 13 | -0.967 | -0.142 | 1.046 | 0.372 | -1.875 | -1.896 | -1.258 | -0.34 | -1.003 | -0.46 | 0.681 | -0.348 | -1.354 | -0.589 | 0.994 | 0.672 |
| 14 | -1.232 | -0.699 | -3.199 | -1.111 | -1.39 | 0.204 | -2.635 | -0.42 | -0.356 | 1.837 | -1.879 | -0.391 | -1.004 | 1.196 | -2.247 | 0.046 |
| 15 | -1.388 | 0.068 | -1.288 | -4.4 | -1.419 | 2.345 | -4.4 | -0.479 | -3.274 | -2.069 | -1.638 | -4.4 | -1.635 | 0.569 | -0.764 | -1.268 |
| 16 | -1.832 | -1.18 | -1.535 | -3.179 | 1.953 | 1.571 | -1.387 | 0.258 | -1.35 | -1.235 | -2.158 | -1.263 | -2.643 | -0.744 | -1.805 | -1.538 |
| 17 | 1.0 | 0.181 | 1.093 | -0.449 | 1.285 | -0.759 | -1.163 | 0.398 | -1.206 | -1.734 | -1.604 | -2.685 | -1.65 | -1.538 | -0.016 | -0.815 |
| 18 | 0.258 | -0.236 | 1.228 | 0.284 | -0.352 | -0.63 | -1.973 | -0.382 | 0.301 | 0.201 | -0.017 | -0.081 | -0.842 | -0.355 | 0.156 | -1.089 |